Presentation on M. Hillenmeyer at el. intersting paper
advertisement

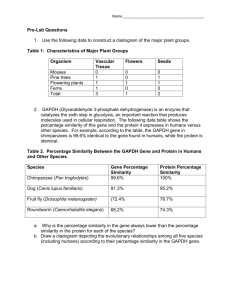
Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. M. Hillenmeyer (Stanford), E. Ericson (Toronto), R. Davis (Stanford), C. Nislow (Toronto), D. Koller (Stanford) and G. Giaever (Toronto) Published in Genome Biology 2010 Presented By: Yaron Margalit 1 • Deeply investigating and analysis chemical genome wide fitness data. – Predict gene-functional – Predict protein-drug interactions – Have new observations or/and extend previous ones with the new data. 2 Outline • Brief introduction • Large-scale genome-wide Dataset • Co-fitness – Motivation and Definition – Implementation – Results • Co-inhibition – Motivation and Definition – Implementation – Results • Predict drug-target interactions – Motivation – Model – Results • Summary 3 Outline • Brief introduction • Large-scale genome-wide Dataset • Co-fitness – Motivation and Definition – Implementation – Results • Co-inhibition – Motivation and Definition – Implementation – Results • Predict drug-target interactions – Motivation – Model – Results • Summary 4 Brief Introduction - Reminder • Deletion Mutants Sensitive to a Particular Drug Should be Synthetically Lethal with the Drug Target Synthetic Lethal Interactions Synthetic Chemical Interactions Alive Alive Drug Alive Alive Drug Dead Dead 5 CGI (C for chemical) vs. GI GI genes Library genes CGI chemicals 6 CGI notes • Some notes we need to take into account when we get into CGI: – Inactivation of the target protein function caused by the compound is not complete – Multi-drug resistant genes: Some mutant are hypersensitive to many drugs of different types (many promiscuous) – Side effects: compound cause inactivation of other proteins and not only the specific gene required 7 Outline • Brief introduction • Large-scale genome-wide Dataset • Co-fitness – Motivation and Definition – Implementation – Results • Co-inhibition – Motivation and Definition – Implementation – Results • Predict drug-target interactions – Motivation – Model – Results • Summary 8 Hillenmeyer et al. Science 2008 Chemical genomic • Study relationship between small molecules and genes. • Small molecules: – Drugs – FDA approved – Chemical probes – well characterized – New compounds – unknown biological activity 10 Saccharomyces cerevisiae (the “beer yeast”) • “Beer yeast” consist of ~ 6000 genes. • ~ 1000 genes are essential • Dataset include large diploid deletion collections – ~ 6000 heterozygous gene deletion strains (+/-) – ~ 5000 homozygous deletion strains (-/-) – Only 5000 because about 1000 are essential (genes that a cell cannot live without regardless of conditions it grows in) 11 Data source • Used deletion sets to study cell growth rate (fitness) response to conditions (small compounds and environmental stressors): – 726 conditions per heterozygous deletion strain – 418 conditions per homozygous deletion strain • Homozygous or heterozygous gene mutation in combination with a drug (or other treatment) causes growth fitness defect (reduction) – Compared to no-drug control 12 Outline • Brief introduction • Large-scale genome-wide Dataset • Co-fitness – Motivation and Definition – Implementation – Results • Co-inhibition – Motivation and Definition – Implementation – Results • Predict drug-target interactions – Motivation – Model – Results • Summary 14 co-fitness • Definition: co-fitness value - the similarity of two genes fitness score across experiments • Intuitive: – Gene-drug interaction: retrieve fitness defect score: compare gene’s intensity in a specific treatment to the same gene’s intensity in the control (no-drug) – Result to gene-gene relationship: Calculate correlation (similarity) between two genes (i.e. “how much genes are sensitive to similar drugs”) • co-fitness was calculated separately for the heterozygous and homozygous datasets 15 co-fitness – the similarity of two genes • How to calculate fitness defect (reduction) gene-drug interaction: – – – – Z-score P-value Log ratio Log P-value • Example of such a score, log rate: Where: - mean intensity of i replicate across multiple control conditions (controls) - intensity of i replicate under treatment t (cases) 16 co-fitness – the similarity of two genes • Calculate correlation gene-gene relationship. • Example of co-fitness, distance metric: Euclidean distance: Where: - i replicate, defect score of gene x under treatment t - i replicate, defect score of gene y under treatment t 17 co-fitness – the similarity of two genes • Goal: Quantify the degree to which co-fitness can predict gene function and compare its performance to other similarities types (datasets) • Several similarities – correlation based were tested: – Pearson correlation – Spearman rank correlation – Euclidean distance – Bicluster cooccurrence count – Bicluster Pearson correlation 18 co-fitness – picking best distance metric 19 co-fitness – the similarity of two genes • So far: We tested and found that Pearson correlation exhibit the best performance for co-fitness • Use co-fitness and evaluate its prediction of gene functional 20 co-fitness predicts reference network • Evaluate co-fitness prediction on expertcurated reference interaction (“reference network”) – gold standard compared dataset. • Each dataset compared to the reference network: – Reference network divided into 32 GO slim biological sub-net works – Each gene pair was assigned to the sub-network if both genes were annotated to that process 21 co-fitness predicts reference network 22 23 24 co-fitness more results • Essential genes were co-fit with other essential genes more frequently: – 40% essential genes co-fit with essential genes compared to 23% for non essential genes. • Pairs of co-complexed genes (genes encoded within same protien complex) increased cofitness with other members of the complex. 25 co-fitness more results 26 co-fitness application example • Find nonessential proteins that might be essential for optimal growth in conditions. – Idea comes from previous study saying proteins that are essential in rich medium (type of condition) tend to cluster into complexes (i.e. essential complex). • Application: – Define complex to be essential if 80% of its members are essential. – Run over all co-fitness values and search for a significant essential complexes. 27 co-fitness application example • Create a synthetic data for each condition: – Generate 10,000 a random distribution – reassign genes to complexes (but maintain complexes size) – Protein complex is essential if at least 80% of its genes had a significant (P < 0.01 cutoff) fitness defect. • Identify condition with significantly more essential complexes if this essential complex was not observed essential in any of the 10,000 permutations. 28 Outline • Brief introduction • Large-scale genome-wide Dataset • Co-fitness – Motivation and Definition – Implementation – Results • Co-inhibition – Motivation and Definition – Implementation – Results • Predict drug-target interactions – Motivation – Model – Results • Summary 29 Co-inhibition • Definition: co-inhibition value: correlation between drug1 and drug2 s.t. inhibit similar genes. • Intuitive (similar to co-fitness): – Gene-drug interaction: retrieve fitness defect score: compare gene’s intensity in a specific treatment to the same gene’s intensity in the control (no-drug) – Result to drug-drug relationship: Calculate correlation (similarity) between two drugs (i.e. “how much drugs inhibit similar genes”) • co-inhibition was calculated separately for the heterozygous and homozygous datasets 30 Co-inhibition • Claim that indicated from small scale databases: High co-inhibition value tend to share chemical structure and mechanism of action in the cell • Goal: use co-inhibition to predict mechanism of action and therefore identify drug targets or toxicities • Next steps: – – – – Calculate co-inhibition (1) Define chemical structural similarity (2) Define chemical therapeutic (action) use (3) Verify claim (1,2,3 share high percent similarity) 31 Co-inhibition • Claim that indicated from small scale databases: High co-inhibition value tend to share chemical structure and mechanism of action in the cell • Goal: use co-inhibition to predict mechanism of action and therefore identify drug targets or toxicities • Next steps: – – – – Calculate co-inhibition (1) Define chemical structural similarity (2) Define chemical therapeutic (action) use (3) Verify claim (1,2,3 share high percent similarity) 32 Calculate co-inhibition (1) • How to calculate fitness defect (reduction) gene-drug interaction – Similar to co-fitness – – – – Z-score P-value Log ratio Log P-value • Example of such a score, log rate: Where: - mean intensity of i replicate across multiple control conditions (controls) - intensity of i replicate under treatment t (cases) 33 Calculate co-inhibition (1) • Calculate correlation drug-drug relationship. • co-inhibition, distance metric that was used Pearson correlation: Where: - i replicate, defect score of drug x with gene g - i replicate, defect score of drug y with gene g 34 Co-inhibition • Claim that indicated from small scale databases: High co-inhibition value tend to share chemical structure and mechanism of action in the cell • Goal: use co-inhibition to predict mechanism of action and therefore identify drug targets or toxicities • Next steps: – – – – Calculate co-inhibition (1) Define chemical structural similarity (2) Define chemical therapeutic (action) use (3) Verify claim (1,2,3 share high percent similarity) 35 Define chemical structural similarity (2) • Model each chemical to substructure motifs • Construct substructure vectors (containing all possible substructures in our case 554 types) and set a value between 0-1 for each substructure is it similar to chemical structure or not. • Calculate structural similarity between 2 drugs by a distance metric. 36 Define chemical structural similarity (2) • Model each chemical to substructure motifs • Construct substructure vectors (containing all possible substructures in our case 554 types) and set a value between 0-1 for each substructure is it similar to chemical structure or not. • Calculate structural similarity between 2 drugs by a distance metric. 37 Define chemical structural similarity (2) • Construct substructure vectors (containing all possible substructures in our case 554 types) and set a value between 0-1 for each substructure is it similar to chemical structure or not. – We will show 3 different ways to do that 38 chemical structural similarity – substructure vectors • First way Binary identifier • Simple binary vector where the value is 1 if the compound contains the substructure and 0 otherwise. 39 chemical structural similarity – substructure vectors • Second way IDF • Convert binary indicator to an inverse document frequency (IDF). IDF score for substructure mofit i (regardless of the chemical): C – number of compounds Cj – number of compounts that contain motif i • Set 0 if compound does not contain substructure and IDF > 0 otherwise. 40 chemical structural similarity – substructure vectors • Third way Binary-IDF • Convert binary indicator to an inverse document frequency (IDF). • Convert back to binary using a threshold on IDF value (for IDF > X threshold set 1 otherwise 0) 41 Define chemical structural similarity (2) • Model each chemical to substructure motifs • Construct substructure vectors (containing all possible substructures in our case 554 types) and set a value between 0-1 for each substructure is it similar to chemical structure or not. • Calculate structural similarity between 2 drugs by a distance metric. 42 Calculate chemical structural similarity (2) • For the binary data (first and third ways) they tested as a distance metric: – Tanimoto (Jaccard) coefficient – Hamming distance – Dice coefficient • For the IDF data (second way) they tested: – – – – – Cosine distance Pearson correlation Spearman correlation Euclidean distance Kendall’s Tau City-block distance 43 Calculate chemical structural similarity (2) • Greatest relationship done by using Binary-IDF with (threshold > 2.5) • Distance metric was Tanimoto (Jaccard) coefficient • Suggests that structure similarity should be defined by a less common substructures. 44 Co-inhibition • Claim that indicated from small scale databases: High co-inhibition value tend to share chemical structure and mechanism of action in the cell • Goal: use co-inhibition to predict mechanism of action and therefore identify drug targets or toxicities • Next steps: – – – – Calculate co-inhibition (1) Define chemical structural similarity (2) Define chemical therapeutic (action) use (3) Verify claim (1,2,3 share high percent similarity) 45 Define chemical therapeutic (action) use (3) • Use known data: – Define pair of compounds to be co-therapeutic if they share annotation at level 3 of the WHO (classification of drug uses) ATC hierarchy. 46 Co-inhibition • Claim that indicated from small scale databases: High co-inhibition value tend to share chemical structure and mechanism of action in the cell • Goal: use co-inhibition to predict mechanism of action and therefore identify drug targets or toxicities • Next steps: – – – – Calculate co-inhibition (1) Define chemical structural similarity (2) Define chemical therapeutic (action) use (3) Verify claim (1,2,3 share high percent similarity) 47 co-inhibition - is it really true? • Counted pairs of compounds that have: – Positive co-inhibition (correlation > 0) – Shared therapeutic class – Measurable structural similarity • From this counting: – 70% did not share structural similarity (Tanimoto similarity < 0.2) 48 co-inhibition – results • Limited correlation between co-inhibition and similar chemical structure. 49 co-inhibition – results • Significant relationship between shared ATC therapeutic class and co-fitness 50 co-inhibition – results • Some observation of differences between shared structure and common therapeutic 51 co-inhibition – results • Co-inhibition can reveal both shared structure and common therapeutic • specially useful for the non target drug use 52 Outline • Brief introduction • Large-scale genome-wide Dataset • Co-fitness – Motivation and Definition – Implementation – Results • Co-inhibition – Motivation and Definition – Implementation – Results • Predict drug-target interactions – Motivation – Model – Results • Summary 53 Predict drug-target interactions • Method to address the difficult task of predicting drug targets. • Goal: – Use genomic data to better predict the protein target of a compound – Distinguish which of the sensitive genes is most likely drug target • Let’s use a Machine-learning algorithm! 54 What is Machine learning • Automated learning. • There are many types of machine learning, we will focus on Supervised, Batch learning (our case). – “Supervised” : Based Training set so that learner should figure out a rule for new arrival data. – “Batch” : Retrieve first training set then run on test set. 55 Machine learning example • Papayas example 56 Predict drug-target interactions • Method to address the difficult task of predicting drug targets. • Learn to estimate an “interaction score” between compound c and gene g: – Have a training set – Set several key features – Produce an estimation for compound c and gene g – Test algorithms using “cross-validation” 57 Predict drug-target interactions • Method to address the difficult task of predicting drug targets (protein-compund interaction). • Learn to estimate an “interaction score” between compound c and gene g: – Have a training set – Set several key features – Produce an estimation for compound c and gene g – Test algorithms using “cross-validation” 58 Training set (1) • Experts identify known protein interactions in yeast (with literature evidence) – 83 training data • In order to test our learning algorithm, have a negative test set of 83 random combinations of compound-protein interactions. 59 Training set (2) • Use known dataset DrugBank for Humans and map it to yeast by application BLASTp. – 46 training data • Again another negative test set of 46 random combinations 60 Predict drug-target interactions • Method to address the difficult task of predicting drug targets. • Learn to estimate an “interaction score” between compound c and gene g: – Have a training set – Set several key features – Produce an estimation for compound c and gene g – Test algorithms using “cross-validation” 61 Key features • Features used in learning drug targets over all 20 features: – Fitness defect score of the heterozygous data (two features) • Log ratio • P-value – Gene sensitivity frequency (one feature) • Number of compounds causing sensitivity in protein. – Drug inhibition frequency (one feature) • Number of inhibit genes. 62 Key features (2) • Features used in learning drug targets over all 20 features: – Phenotype in rich medium (one feature) – Chemical structure similarity enrichment of putative compounds (three features) • Sensitive gene for similar compounds might increase confidence • Number of other compounds that share a common motif with the requested compound • Average structural similarity score 63 Key features (3) • Features used in learning drug targets over all 20 features: – Co-inhibition “secondary compound” fitness defect scores (ten features) • Top 10 co-inhibiting compounds with the requested compound – Co-inhibition “secondary compound” summary statistics fitness defect scores (two features): • Mean • Median 64 Machine learning algorithm • Several machine learning algorithms were used: – Random forest – Naïve Bayes – Decision Stump – Logistic regression – SVM – Decision tree – Bayesian Network 65 Machine learning validation • 10-fold Cross-validation method: – Partition the training set into 10 subsets – For each subset, a predictor is trained on the other 9 subsets and then its error is estimated using the subset. – Pick algorithm with minimal errors. 66 Random forest is the best algorithm 67 Random Forest is really useful? Why not just use fitness defect score? 68 69 Intro to decision tree 70 From decision tree to Random Forest • Forest = Multiple decision trees – The output of every decision tree in the “forest” is averaged • What’s random in a Random Forest? – Random a subset of the explanatory variables – Random a subset of the training data • Why random? – Avoids modeling noise – Decision trees are greedy: Using the best split at every point might overlook better solutions in the long-term (stuck at local optimum) 71 Why random forests are great • Non parametric and non-linear: – No specific relationship between our explanatory variables and our predictions. – Logistic regression (other algorithm) would impose for example a specific relationship between the explanatory variables and the predicated value. – Random forest is flexible. No need for special assumptions or specific decisions. All decision are random. – Another advantage: incorporate interactions between all the explanatory variables. 72 Random forest algorithm • Each tree: – Take number of training cases and number of variables (key features) – Calculate the best split cases on these variables. – Each tree is grown until the end (full tree) • Prediction: – Each label assigned to a value according to each tree. – Take the average vote. 73 Prediction results • Authors run algorithm over the genome-wide dataset • 4 of top 10 predicated interactions were validated in lab 74 Summary • We have shown a systematic analysis for genome-wide large scale fitness data. – Introduced co-fitness value for gene-gene relationship . Helpful to predict gene functionality – Defined similar drug relationship by co-inhibition value. Helpful to show chemical similar structure and therapeutic use. – Showed a learning algorithm to predict drugtargets 75 Questions 76 77 Calculate co-fitness • Pearson correlation: Where: - i replicate, defect score of gene x under condition g - i replicate, defect score of gene y under condition g 78