SITC2015 - Genocea Biosciences
advertisement
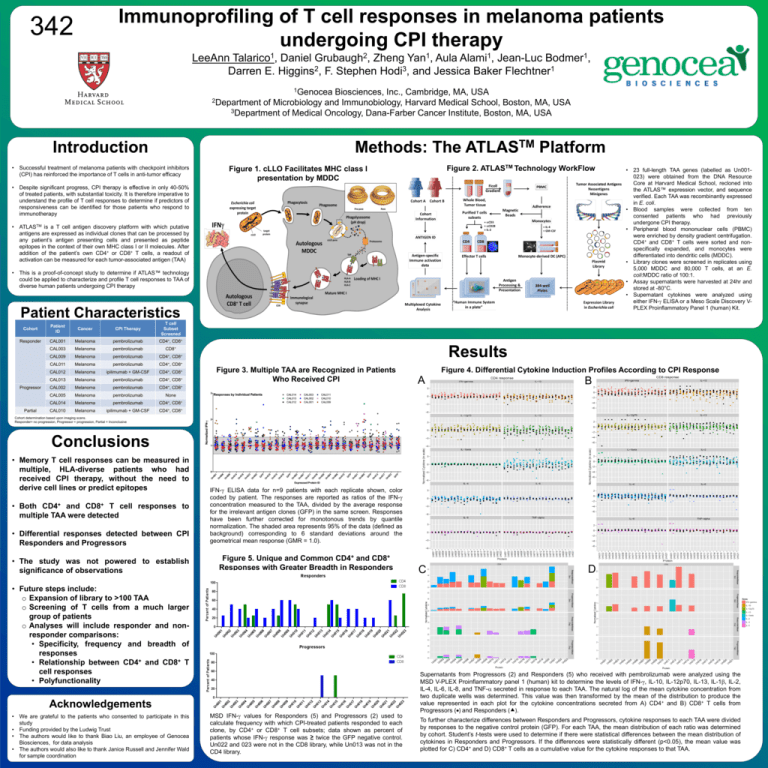
Immunoprofiling of T cell responses in melanoma patients undergoing CPI therapy 342 LeeAnn Talarico1, Daniel Grubaugh2, Zheng Yan1, Aula Alami1, Jean-Luc Bodmer1, Darren E. Higgins2, F. Stephen Hodi3, and Jessica Baker Flechtner1 1Genocea Biosciences, Inc., Cambridge, MA, USA 2Department of Microbiology and Immunobiology, Harvard Medical School, Boston, MA, USA 3Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA Introduction Methods: The • Successful treatment of melanoma patients with checkpoint inhibitors (CPI) has reinforced the importance of T cells in anti-tumor efficacy TM ATLAS Platform Figure 2. ATLASTM Technology WorkFlow Figure 1. cLLO Facilitates MHC class I presentation by MDDC • Despite significant progress, CPI therapy is effective in only 40-50% of treated patients, with substantial toxicity. It is therefore imperative to understand the profile of T cell responses to determine if predictors of responsiveness can be identified for those patients who respond to immunotherapy • ATLASTM is a T cell antigen discovery platform with which putative antigens are expressed as individual clones that can be processed by any patient’s antigen presenting cells and presented as peptide epitopes in the context of their own MHC class I or II molecules. After addition of the patient’s own CD4+ or CD8+ T cells, a readout of activation can be measured for each tumor-associated antigen (TAA) • This is a proof-of-concept study to determine if ATLAS™ technology could be applied to characterize and profile T cell responses to TAA of diverse human patients undergoing CPI therapy Patient Characteristics Cohort Patient ID Cancer CPI Therapy T cell Subset Screened Responder CAL001 Melanoma pembrolizumab CD4+, CD8+ CAL003 Melanoma pembrolizumab CD8+ CAL009 Melanoma pembrolizumab CD4+, CD8+ CAL011 Melanoma pembrolizumab CD4+, CD8+ CAL012 Melanoma ipilimumab + GM-CSF CD4+, CD8+ CAL013 Melanoma pembrolizumab CD4+, CD8+ CD4+, Progressor Partial Results Figure 3. Multiple TAA are Recognized in Patients Who Received CPI CD8+ CAL002 Melanoma pembrolizumab CAL005 Melanoma pembrolizumab None CAL014 Melanoma pembrolizumab CD4+, CD8+ CAL010 Melanoma ipilimumab + GM-CSF CD4+, CD8+ • 23 full-length TAA genes (labelled as Un001023) were obtained from the DNA Resource Core at Harvard Medical School, recloned into the ATLAS™ expression vector, and sequence verified. Each TAA was recombinantly expressed in E. coli. • Blood samples were collected from ten consented patients who had previously undergone CPI therapy. • Peripheral blood mononuclear cells (PBMC) were enriched by density gradient centrifugation. CD4+ and CD8+ T cells were sorted and nonspecifically expanded, and monocytes were differentiated into dendritic cells (MDDC). • Library clones were screened in replicates using 5,000 MDDC and 80,000 T cells, at an E. coli:MDDC ratio of 100:1. • Assay supernatants were harvested at 24hr and stored at -80°C. • Supernatant cytokines were analyzed using either IFN-g ELISA or a Meso Scale Discovery VPLEX Proinflammatory Panel 1 (human) Kit. Figure 4. Differential Cytokine Induction Profiles According to CPI Response A B C D Cohort determination based upon imaging scans. Responder= no progression, Progressor = progression, Partial = Inconclusive Conclusions • Memory T cell responses can be measured in multiple, HLA-diverse patients who had received CPI therapy, without the need to derive cell lines or predict epitopes • Both CD4+ and CD8+ T cell responses to multiple TAA were detected • Differential responses detected between CPI Responders and Progressors • The study was not powered to establish significance of observations IFN-g ELISA data for n=9 patients with each replicate shown, color coded by patient. The responses are reported as ratios of the IFN-g concentration measured to the TAA, divided by the average response for the irrelevant antigen clones (GFP) in the same screen. Responses have been further corrected for monotonous trends by quantile normalization. The shaded area represents 95% of the data (defined as background) corresponding to 6 standard deviations around the geometrical mean response (GMR = 1.0). Figure 5. Unique and Common CD4+ and CD8+ Responses with Greater Breadth in Responders • Future steps include: o Expansion of library to >100 TAA o Screening of T cells from a much larger group of patients o Analyses will include responder and nonresponder comparisons: • Specificity, frequency and breadth of responses • Relationship between CD4+ and CD8+ T cell responses • Polyfunctionality Acknowledgements • We are grateful to the patients who consented to participate in this study • Funding provided by the Ludwig Trust • The authors would like to thank Biao Liu, an employee of Genocea Biosciences, for data analysis • The authors would also like to thank Janice Russell and Jennifer Wald for sample coordination MSD IFN-g values for Responders (5) and Progressors (2) used to calculate frequency with which CPI-treated patients responded to each clone, by CD4+ or CD8+ T cell subsets; data shown as percent of patients whose IFN-g response was ≥ twice the GFP negative control. Un022 and 023 were not in the CD8 library, while Un013 was not in the CD4 library. Supernatants from Progressors (2) and Responders (5) who received with pembrolizumab were analyzed using the MSD V-PLEX Proinflammatory panel 1 (human) kit to determine the levels of IFN-g, IL-10, IL-12p70, IL-13, IL-1b, IL-2, IL-4, IL-6, IL-8, and TNF-a secreted in response to each TAA. The natural log of the mean cytokine concentration from two duplicate wells was determined. This value was then transformed by the mean of the distribution to produce the value represented in each plot for the cytokine concentrations secreted from A) CD4+ and B) CD8+ T cells from Progressors () and Responders (). To further characterize differences between Responders and Progressors, cytokine responses to each TAA were divided by responses to the negative control protein (GFP). For each TAA, the mean distribution of each ratio was determined by cohort. Student’s t-tests were used to determine if there were statistical differences between the mean distribution of cytokines in Responders and Progressors. If the differences were statistically different (p<0.05), the mean value was plotted for C) CD4+ and D) CD8+ T cells as a cumulative value for the cytokine responses to that TAA. www.postersession.com




