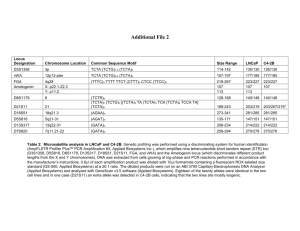
condon_02_13_03
advertisement
Computational Prediction of RNA and DNA Secondary Structure Anne Condon Bioinformatics, and Empirical and Theoretical Algorithmics (BETA) Laboratory The Department of Computer Science, UBC RNA plays varied roles in the cell… “… a familiar performer has turned up in a stunning variety of guises. RNA, long upstaged by its more glamorous sibling, is turning out to have star qualities of its own” – J. Couzin, Science, 2003 … in sequence analysis, cDNA to be sequenced TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA TTAC AATC TACT ATCA ACAT TCTA CTTT CAAA address tag ... ... ... ... “…signature sequencing [permits] application of powerful statistical techniques for discovery of functional relationships among genes” – S. Brenner et al. …nanotechnology, ACCAGGT U R. Hamers “Linking biological molecules with nanotubes and nano wires [is] a method for biological sensing and for controlling nanoscale assembly” – R. Hamers, Chemistry, U. Wisconsin and beyond… “..rather than examining in detail what occurs in nature (biological organisms), we take the engineering approach of asking, "what can we build?" E. Winfree … hold promise in therapeutics, and hold clues to our understanding of primitive life “there are strong reasons to conclude that DNA and protein based life was preceded by a simpler life form based primarily on RNA” – G. F. Joyce To understand the function of RNA molecules, we need to understand their structure. DNA, RNA secondary structure 5' 3' our goals • predict from base sequence the secondary structure of – a single molecule – a small group of molecules – the most stable molecule in a combinatorial set of molecules • design a molecule or molecules with a certain secondary structure overview • background – how is RNA secondary structure prediction currently done? • challenges – how might it be done better? • projects – what is the BETA-lab doing about it? approaches to RNA secondary structure prediction • comparative sequence analysis • prediction from base sequence – find minimum free energy (mfe) structure free energy model • free energy of structure (at fixed temperature, ionic concentration) = sum of loop energies • standard model uses experimentally determined thermodynamic parameters where available; extrapolations for long loops free energy model • free energy of structure (at fixed temperature, ionic concentration) = sum of loop energies • standard model uses experimentally determined thermodynamic parameters where available; extrapolations for long loops on the mfe approach • mfe approach ignores folding pathway, metal ions, nonstandard bonds • “some species can remain kinetically trapped in nonequilibrium states… we expect that most RNA’s exist naturally in their thermodynamically most stable configurations” –Tinoco and Bustamante, J. Mol. Biol. 1999. why is mfe secondary structure prediction hard? • mfe structure can be found by calculating free energy of all possible structures • but, number of potential structures grows exponentially with the number, n, of bases • structures can be arbitrarily complex • success for restricted classes of structures predicting mfe pseudoknot free structures • dynamic programming avoids explicit enumeration of all pseudoknot free structures (Zuker & Stiegler 1981) • suboptimal folds, probabilities of base pairings can also be calculated • software: mfold, Vienna package dynamic programming (Zuker and Steigler) • based on the “more is less” principle: by calculating more than you need, less work is needed overall • construct mfe structure for whole strand from mfe structures for substrands • running time is O(n3) dynamic programming (Zuker and Steigler) • W(i,j): mfe structure of substrand from i to j • V(i,j): mfe structure of substrand from i to j, in which ith and jth bases are paired V(i,j) W(i,j) i j i j recurrences = W(i,j) i j min V(i,j) i W(i,k) j i W(k+1,j) k k+1 j recurrences = i j min i j i i+1 k k+1 j-1 j i k l j recurrences = i j = i min j i j i k k+1 j i j i i+1 k k+1 j-1 j i k l min j current approaches: pseudoknotted structures • loop energies augmented to fit known data • mfe algorithms have been extended to handle certain pseudoknotted structures (Akutsu, 2000; Rivas and Eddy, 1999) • heuristic approaches also used (STAR software, van Batenburg et al.) W(i,j): Rivas and Eddy algorithm = min i j i j i k k+1 j i r k l j recurrence for gapped structures = min i k ir i j l k k r l j l j ir k l j i k l s j s i k i l k l s j j Rivas and Eddy algorithm • running time is O(n6) • “we lack a systematic a priori characterization of the class of configurations that this algorithm can solve” (Rivas and Eddy, 1999) heuristic approaches • space of structures is explored, guided by random decisions as well as goal of minimizing free energy • can incorporate better energy models, model folding pathways, and be substantially faster than mfe algorithms • no guarantee that optimal solution is found STAR algorithm (van Batenburg et al.) • simulates RNA folding by stepwise addition and removal of stems to the structure formed at previous steps • choice of stem to add/remove is random, biased by “fitness” of resulting structure • process is carried out on large population of structures, not just one prediction challenges • predict structures formed from multiple strands • predict which strand in a combinatorial set has minimum free energy structure • predict pseudoknotted structures BETA-Lab Projects • predict structures formed from multiple strands PairFold • predict which strand in a combinatorial set has minimum free energy structure CombFold • predict pseudoknotted structures PseudoSpotter PairFold (Andronescu) • predicts minimum energy pseudoknot free secondary structure of a pair of strands • energy model: loop plus initiation energies from Turner, Santa Lucia labs CombFold (Andronescu et al.) • predicts which strand in a combinatorial set has minimum free energy structure TTAC TTAC TTAC TTAC TTAC TTAC TTAC TTAC AATC AATC AATC AATC AATC AATC AATC AATC TACT TACT TACT TACT TACT TACT TACT TACT ATCA ATCA ATCA ATCA ATCA ATCA ATCA ATCA ACAT ACAT ACAT ACAT ACAT ACAT ACAT ACAT TCTA TCTA TCTA TCTA TCTA TCTA TCTA TCTA CTTT CTTT CTTT CTTT CTTT CTTT CTTT CTTT CAAA CAAA CAAA CAAA CAAA CAAA CAAA CAAA • uses dynamic programming to avoid examining all combinatorial possibilities CombFold (Andronescu et al.) • predicts which strand in a combinatorial set has minimum free energy structure TTAC TTAC TTAC TTAC TTAC TTAC TTAC TTAC AATC AATC AATC AATC AATC AATC AATC AATC TACT TACT TACT TACT TACT TACT TACT TACT ATCA ATCA ATCA ATCA ATCA ATCA ATCA ATCA ACAT ACAT ACAT ACAT ACAT ACAT ACAT ACAT TCTA TCTA TCTA TCTA TCTA TCTA TCTA TCTA CTTT CTTT CTTT CTTT CTTT CTTT CTTT CTTT CAAA CAAA CAAA CAAA CAAA CAAA CAAA CAAA • TCTAATCATACTAATCCTTTTACTATCATTAC has a slightly stable secondary structure at 37oC PseudoSpotter (Ren et al. ) • heuristic algorithm for predicting pseudoknotted structures PseudoSpotter (Ren et al. ) PseudoSpotter (Ren et al. ) 1 P4 P1 5’ 3’ PseudoSpotter (Ren et al. ) 1 P4 P1 5’ 3’ 2 5’ 3’ P2 PseudoSpotter (Ren et al. ) 1 2 P4 P1 5’ 3’ P2 5’ 3’ 3 5’ P3 comparison of pseudoknot algorithms R&E STAR Pseudospotter Optimal Subopt, Fail Close 18 5 9 19 13 21 7 4 • Quality of solutions found over 32 sample strands taken from RDB, Pseudobase web page next steps • better energy parameters for model, model evaluation and refinement • develop better heuristic algorithms for predicting pseudoknotted structures • improved mfe algorithms for natural pseudoknotted structures • suboptimal foldings, partition function how to get better energy models? • compare variations of standard free energy model in terms of prediction quality • perform lab experiments to find energy parameters • incorporate tertiary structural elements (nonstandard base pairs or motifs, folding pathway, metal ions) how to improve heuristic algorithm? 1 P4 P1 5’ 2 3’ 5’ 3’ P2 maybe here? how to improve mfe algorithm? • we have a simple characterization of the class of structures that the Rivas and Eddy algorithm can handle • perhaps an O(n5) algorithm for natural structures? acknowledgements Dr. Holger Hoos BETA-lab co-founder Rosalia Aguirrez-Hernandez RNA design Mirela Andronescu Pairfold, Combfold Viann Chan XIST RNA structure Jihong Ren Pseudospotter Sohrab Shah structural motif finding Dan Tulpan DNA word design Shelly Zhao Combfold, RNA structure analysis Drs. Rob Corn, Lloyd Smith University of Wisconsin Bioinformatics, and Empirical and Theoretical Algorithmics (BETA) Laboratory www.cs.ubc.ca/labs/beta