Lecture 3
advertisement
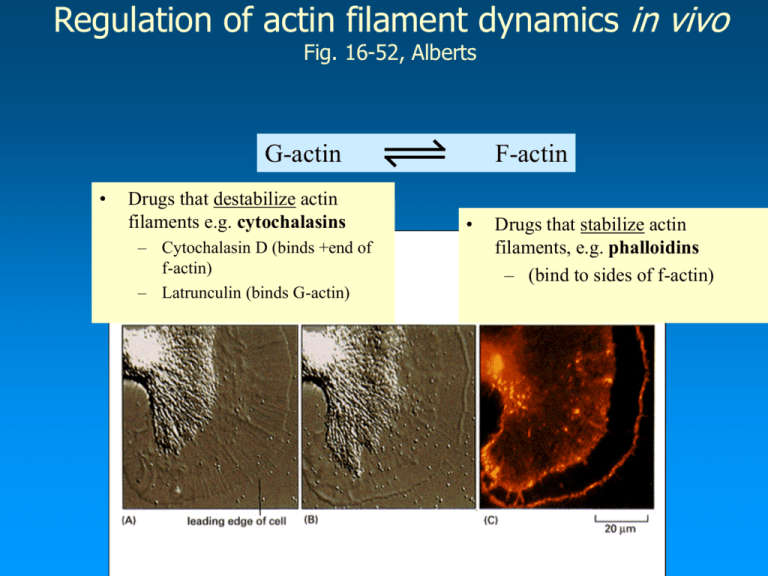
Regulation of actin filament dynamics in vivo Fig. 16-52, Alberts G-actin • Drugs that destabilize actin filaments e.g. cytochalasins – Cytochalasin D (binds +end of f-actin) – Latrunculin (binds G-actin) F-actin • Drugs that stabilize actin filaments, e.g. phalloidins – (bind to sides of f-actin) The Roles of Actin Binding Proteins Dynamics/structure Structure: Filamin, ABP-120, 240 etc., Structure:Fimbrin, Villin, -actinin Actin-Membrane Binding: Vinculin, Ponticulin, -actinin Dynamics:Cofilin, ADF Dynamics:Gelsolin Actin binding proteins that regulate dynamics Lodish 5th Ed. Chapter 19, p786-791 • Actin binding proteins (APB’s) modulate the function of the actin cytoskeleton • 1. Dynamics • Thymosin -4 (G-actin sequesterer) • Profilin (Increases rate of polymerization) • Gelsolin (Increases rate of actin filament turnover) • Capping proteins (Increases rate of polymerization) • Arp2/3 (Nucleation ) Thymosin -4 and Profilin are monomer sequestering proteins • Fact: The Cc for actin filament polymerization is 0.1uM, the total actin concentration in a cell is 200uM. 40% of actin in cells is unpolymerized. Why ? • Proteins in the cytoplasm sequester or bind to actin monomers - preventing them from polymerizing. Factors which influence the binding of these proteins to actin monomers will affect the rate of actin polymerization. Active sequesterers Inactive sequesterers Microinjection of excess TB4 into cells causes loss of stress fibers • • • Although actin stress fibers are relatively stable turnover of actin monomers is occurring. Monomers leaving a stress fiber will be rapidly sequestered by TB4. Gradually the stress fiber will disappear. The equilibrium is shifted toward increasing monomer concentration at the expense of f-actin. After Before The 3D structure of the actin-profilin complex Fig. 18-14 • Profilin is a weak actin sequesterer • It binds opposite the ATP binding cleft (in contrast to T-4) • This allows exchange of ADP for ATP to occur (recharging the monomer) • Profilin facilitates addition of actin monomers to the growing + end of Factin – Because affinity of actin-profilin complex > than single actin monomer – it speeds up rate of polymerization at +end Regulation of actin polymerization by profilin Fig. 18-15 Regulation of actin polymerization by profilin (explanation of previous slide) • A)Before a signaling event, profilin is inactive because it is bound to PIP2 in the plasma membrane. • B1) Following a signaling event PIP2 is cleaved by phospholipase C, releasing profilin – which is now active. B2) When a small amount of profilin is activated it completes with T-4 for G-actin and rapidly adds it to the +end of F-actin. • • C) In addition to increasing the rate of actin polymerization at the + end, profilin sequesters ADP-G-actin, allowing exchange of ADP for ATP – thus increasing the pool of ATP-G actin (which indirectly increases the rate of actin polymerization). • The activity of profilin is maintained close to the plasma membrane because the activated profilin binds to acidic membrane phospholipids and certain proline rich proteins (will talk about these later) that localize at the plasma membrane. Actin binding proteins modulate the structure of the actin cytoskeleton Fig. 16-65 and 66, Alberts • • Actin forms a wide variety of structures in the cell, some are more permanent than others. Many different structures can coexist in the same cell. There are 3 basic types of actin structure – contractile bundles-containing -actinin, – gel-like networks containing filamin – tight parallel bundles containing fimbrin. filamin -actinin fimbrin Actin filaments and actin-binding proteins in a microvillus • • • • Microvilli are permanent finger-like projections (0.08um wide, 0.5-10um in length) Each structure contains a core of actin filaments cross-linked by proteins such as fimbrin, villin and myosin I (crosslinks F-actin to the plasma membrane) They are found were the cell membrane faces a fluid environment. They increase surface area up to 20 times in gut epithelial cells Actin structures formed by ABP’s Filopodia (fimbrin) Actin meshworks form ruffles on cell surface (filamin) ( -actinin) ABP’s can link actin filaments to the plasma membrane • Function: They attach to actin cortex to integral membrane proteins • Provide stiffness to cell membranes and immobilizes some membrane proteins. Filamin provides rigidity to lamellipodia and support for the plasma membrane • • • • • Cells that lack filamin, cannot move because cells cannot form rigid protrusions Cells form blebs because the actin cortex is not stiff enough to prevent the cell’s cytoplasm from “bulging - out” When the filamin gene is transfected into the same cell type, cells can move and do not form blebs Filamin crosslinks actin filaments into 3-dimensional gels and flat sheets The hinge region of filamin allows actin filaments to change orientation with respect to each other Actin binding proteins that link the plasma membrane to the actin cytoskeleton • The actin filaments within cellular surface features, must be linked to the membrane since the actin cytoskeleton exerts protrusive and contractile forces on it. – e.g. endocytosis, phagocytosis – protrusion and retraction of the cell edge during cell movement ERM proteins attach actin filaments to the plasma membrane Ch. 16, p.936 • • • • ABPs that crosslink the actin cytoskeleton to the plasma membrane are important for cell shape and strength of the plasma membrane Ezrin, radixin, moesin were the first three ERM proteins discovered C-term. domain binds actin, N-term. domain binds transmembrane proteins such as CD44 The activity of these proteins is regulated by extra and intracellular signal – Folded state - inactive, unfolded state, active







