COST Functional Modeling Workshop
advertisement

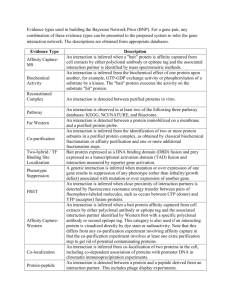
Introduction to the Gene Ontology: A User’s Guide COST Functional Modeling Workshop 22-24 April, Helsinki Introduction to GO • • • • The Gene Ontology Consortium The Gene ontology A GO annotation example GO evidence codes • no GO vs ND • Making Annotations • Multiple annotations - the gene association (ga) file • Sources of GO THE GENE ONTOLOGY CONSORTIUM http://www.geneontology.org/ The GO Consortium provides: • central repository for ontology updates and annotations • central mechanism for changing GO terms (adding, editing, deleting) • quality checking for annotations • consistency checks for how annotations are made by different groups • central source of information for users • co-ordination of annotation effort GO Consortium and GO Groups: • groups decide gene product set to annotate • biocurator training • tool development mostly by groups • many non-consortium groups • education and training by groups • outreach to biocurators/databases by GOC Annotation Strategy • Experimental data • many species have a body of published, experimental data • Detailed, species-specific annotation: ‘depth’ • Requires manual annotation of literature - slow • Computational analysis • • • • Can be automated - faster Gives ‘breadth’ of coverage across the genome Annotations are general Relatively few annotation pipelines Releasing GO Annotations GO annotations are stored at individual databases Sanity checks as data is entered – is all the data required filled in? Databases do quality control (QC) checks and submit to GO GO Consortium runs additional QC and collates annotations Checked annotations are picked up by GO users eg. public databases, genome browsers, array vendors, GO expression analysis tools AgBase Quality Checks & Releases AgBase Biocurators ‘sanity’ check AgBase biocuration interface ‘sanity’ check & GOC QC AgBase database ‘sanity’ check EBI GOA Project ‘sanity’ check: checks to ensure all appropriate information is captured, no obsolete GO:IDs are used, etc. GO analysis tools Microarray developers ‘sanity’ check & GOC QC GO Consortium database UniProt db QuickGO browser GO analysis tools Microarray developers Public databases AmiGO browser GO analysis tools Microarray developers THE GENE ONTOLOGY Gene Ontology (GO) • Not about genes! • Gene products: genes, transcripts, ncRNA, proteins • The GO describes gene product function • Not a single ontology • Biological Process (BP or P) • Molecular Function (MF or F) • Cellular Component (CC or C) • de facto method for functional annotation • Widely used for functional genomics (high throughput). What the GO doesn’t do: • Does not describe individual gene products • e.g. cytochrome c is not in the GO but oxidoreductase activity is • Does not describe mutants or diseases, e.g. oncogenesis. • Does not include sequence attributes, e.g., exons, introns, protein domains. • Is not a database of sequences. What is the Gene Ontology? “a controlled vocabulary that can be applied to all organisms even as knowledge of gene and protein roles in cells is accumulating and changing” • assign functions to gene products at different levels, depending on how much is known about a gene product • is used for a diverse range of species • structured to be queried at different levels, eg: • find all the chicken gene products in the genome that are involved in signal transduction • zoom in on all the receptor tyrosine kinases • human readable GO function has a digital tag to allow computational analysis of large datasets relationships between terms Ontologies digital identifier (computers) description (humans) A GO ANNOTATION EXAMPLE A GO Annotation Example NDUFAB1 (UniProt P52505) Bovine NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa Biological Process (BP or P) GO:0006633 fatty acid biosynthetic process TAS GO:0006120 mitochondrial electron transport, NADH to ubiquinone TAS GO:0008610 lipid biosynthetic process IEA NDUFAB1 GO:0005504 GO:0008137 GO:0016491 GO:0000036 Molecular Function (MF or F) fatty acid binding IDA NADH dehydrogenase (ubiquinone) activity TAS oxidoreductase activity TAS acyl carrier activity IEA Cellular Component (CC or C) GO:0005759 mitochondrial matrix IDA GO:0005747 mitochondrial respiratory chain complex I IDA GO:0005739 mitochondrion IEA A GO Annotation Example NDUFAB1 (UniProt P52505) Bovine NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa GO:ID (unique) aspect or ontology GO evidence code GO term name GO EVIDENCE CODES & MAKING ANNOTATIONS Why record GO evidence code? • GO did not initially record evidence for functional assertion: • NR: Not Recorded • “inferred from…” • deduce or conclude (information) from evidence and reasoning • provides information about the support for associating a gene product with a function • different experiments allow us to draw different conclusions • reliability Types of GO Evidence Codes 1. 2. 3. 4. 5. 6. Experimental Evidence Codes Computational Analysis Evidence Codes Author Statement Evidence Codes Curator Statement Evidence Codes Automatically-assigned Evidence Codes Obsolete Evidence Codes GO EVIDENCE CODES Direct Evidence Codes IDA - inferred from direct assay IEP - inferred from expression pattern IGI - inferred from genetic interaction IMP - inferred from mutant phenotype IPI - inferred from physical interaction Guide to GO Evidence Codes http://www.gene ontology.org/GO.e vidence.shtml Indirect Evidence Codes inferred from literature IGC - inferred from genomic context TAS - traceable author statement NAS - non-traceable author statement IC - inferred by curator inferred by sequence analysis RCA - inferred from reviewed computational analysis IS* - inferred from sequence* IEA - inferred from electronic annotation Other NR - not recorded (historical) ND - no biological data available ISS - inferred from sequence or structural similarity ISA - inferred from sequence alignment ISO - inferred from sequence orthology ISM - inferred from sequence model GO EVIDENCE CODES Direct Evidence Codes GO Mapping IDA - inferred fromExample direct assay IEP - inferred from expression pattern IGI - inferred from genetic interaction IMP - inferred from mutant phenotype IPI - inferred from physical interaction Indirect Evidence Codes inferred from literature IGC - inferred from genomic context TAS - traceable author statement NAS - non-traceable author statement IC - inferred by curator inferred by sequence analysis RCANDUFAB1 - inferred from reviewed computational analysis IS* - inferred from sequence* IEA - inferred from electronic annotation Other NR - not recorded (historical) ND - no biological data available Biocuration of literature • detailed function • “depth” • slower (manual) P05147 Biocuration of Literature: detailed gene function Find a paper about the protein. PMID: 2976880 Read paper to get experimental evidence of function Use most specific term possible experiment assayed kinase activity: use IDA evidence code GO EVIDENCE CODES Direct Evidence Codes GO Mapping IDA - inferred fromExample direct assay IEP - inferred from expression pattern IGI - inferred from genetic interaction IMP - inferred from mutant phenotype IPI - inferred from physical interaction Biocuration of literature • detailed function • “depth” • slower (manual) Indirect Evidence Codes inferred from literature IGC - inferred from genomic context TAS - traceable author statement NAS - non-traceable author statement IC - inferred by curator inferred by sequence analysis RCANDUFAB1 - inferred from reviewed computational analysis IS* - inferred from sequence* IEA - inferred from electronic annotation Other NR - not recorded (historical) ND - no biological data available Sequence analysis • rapid (computational) • “breadth” of coverage • less detailed ISS - inferred from sequence or structural similarity ISA - inferred from sequence alignment ISO - inferred from sequence orthology ISM - inferred from sequence model Computational Analysis Evidence In the beginning: • IGC: Inferred from Genomic Context • e.g. operons • RCA: inferred from Reviewed Computational Analysis • computational analyses that integrate datasets of several types • ISS: Inferred from Sequence or Structural Similarity Computational Analysis Evidence • Then different types of sequence analysis added: ISS: Inferred from Sequence or Structural Similarity • ISO: Inferred from Sequence Orthology • ISA: Inferred from Sequence Alignment • ISM: Inferred from Sequence Model Computational Analysis Evidence • Phylogenetic analysis codes added: • IBA: Inferred from Biological aspect of Ancestor • IBD: Inferred from Biological aspect of Descendant • IKR: Inferred from Key Residues • characterized by the loss of key sequence residues - implies a NOT annotation • IRD: Inferred from Rapid Divergence • characterized by rapid divergence from ancestral sequence – implies a NOT annotation Unknown Function vs No GO • ND – no data • Biocurators have tried to add GO but there is no functional data available • Previously: “process_unknown”, “function_unknown”, “component_unknown” • Now: “biological process”, “molecular function”, “cellular component” • No annotations (including no “ND”): biocurators have not annotated • this is important for your dataset: what % has GO? MULTIPLE ANNOTATIONS: GENE ASSOCIATION FILES The gene association (ga) file • standard file format used to capture GO annotation data • tab-delimited file containing 17* fields of information: • Information about the gene product (database, accession, name, symbol, synonyms, species) • information about the function: • GO ID, ontology, reference, evidence, qualifiers, context (with/from) • data about the functional annotation • date, annotator * GO Annotation File Format 2.0 has two additional columns compared to GAF 1.0: annotation extension (column 16) and gene product form ID (column 17). http://www.geneontology.org/GO.format.gaf-2_0.shtml (additional column added to this example) gene product information metadata: when & who function information Used to give more specific information about the evidence code (not always displayed) Used to qualify the annotation (not always displayed) Gene association files • GO Consortium ga files • many organism specific files • also includes EBI GOA files • EBI GOA ga files • UniProt file contains GO annotation for all species represented in UniProtKB • AgBase ga files • organism specific files • AgBase GOC file – submitted to GO Consortium & EBI GOA • AgBase Community file – GO annotations not yet submitted or not supported / annotations provided by researchers • all files are quality checked http://www.geneontology.org http://www.ebi.ac.uk/GOA/ http://www.agbase.msstate.edu/ Sources of GO 1. Primary sources of GO: from the GO Consortium (GOC) & GOC members • most up to date • most comprehensive 2. Secondary sources: other resources that use GO provided by GOC members • public databases (eg. NCBI, UniProtKB) • genome browsers (eg. Ensembl) • array vendors (eg. Affymetrix) • GO expression analysis tools Sources of GO annotation • Different tools and databases display the GO annotations differently. • Since GO terms are continually changing and GO annotations are continually added, need to know when GO annotations were last updated. Secondary Sources of GO annotation EXAMPLES: public databases (eg. NCBI, UniProtKB) genome browsers (eg. Ensembl) array vendors (eg. Affymetrix) CONSIDERATIONS: What is the original source? When was it last updated? Are evidence codes displayed? Differences in displaying GO annotations: secondary/tertiary sources. For more information about GO • GO Evidence Codes: http://www.geneontology.org/GO.evidence.shtml • gene association file information: http://www.geneontology.org/GO.format.annotation.shtml • tools that use the GO: http://www.geneontology.org/GO.tools.shtml • GO Consortium wiki: http://wiki.geneontology.org/index.php/Main_Page All websites are listed on the AgBase workshop website.