Rhodopsin
advertisement
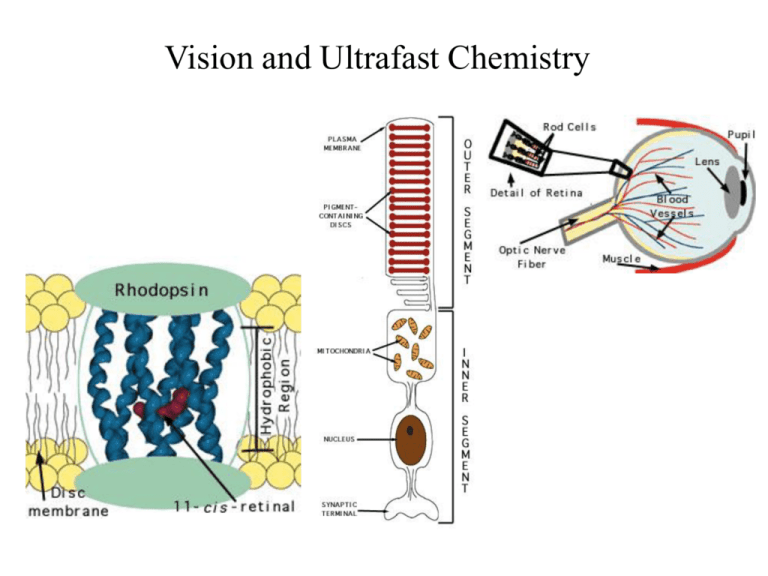
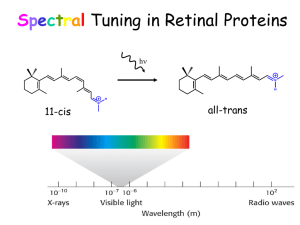
Vision and Ultrafast Chemistry Light Visual signaling Rod G-protein signaling pathway Rhodopsin Cone Visual Receptor Protein Rhodopsin Humphrey et. al., J. Molec. Graphics, 14:33-38, 1996 Freely available, with source code from http://www.ks.uiuc.edu/Research/vmd/ Rhodopsin GPCR, vision in all species Bacteriorhodopsin Photosynthesis, proton pump Organization of the Purple Membrane of Halobacteria Baudry et al, J. Phys. Chem. (in press) hn assembly V Ben-Nun et al Faraday Disc. 110: 447-462 (1998) protein Molnar et al J. Mol. Struct. (in press) molecular electronics H+ function Constructing and Simulating the Purple Membrane assembly function protein Vibrational Spectroscopy (Kyoto) Organic Synthesis (Rehovot) Quantum Chemistry (Heidelberg) Photophysics (Siena) Protein Simulation (Urbana) Pharmacolgy (New York) molecular electronics Molecular Dynamics Program Used: NAMD2 256 192 128 64 0 0 64 128 192 # processors 256 hexagonal unit cell 23700 atoms per unit cell Periodic boundary conditions in 3D (multilayers); NpT (constant pressure) simulations; Particle Mesh Ewald (no electrostatic cutoff); ~2 weeks/ns on 4 Alpha AXP21264-500Mhz procs. Thermodynamics of the Purple Membrane PM thickness NpT simulation: constant temperature, variable volume Reduction of PM thickness during NpT simulation In-plane dimensions Distribution of external water after MD Equilibration of PM: rearrangement of water molecules Before MD After MD water protein “c” dimension perpendicular to the membrane Top view of PM: Water molecules penetrate the PM, but not the protein, stop at Arg82 & Asp96 Asp96 retinal Arg82 Color in Vision cone cells Visual receptors of rhodopsin family are classified based on their color sensitivity Rhodopsin Family of Proteins • Seven transmembrane helices • Retinal chromophore bound to a lysine via the Schiff base Me Me Me N Me Me H protonated Schiff base retinal (PSBR) Color Regulation Visual receptors detect light by electronic excitation of retinal at different wavelengths. 400nm 500nm 600nm Absorption spectra of retinal in different visual receptors Question: How does the protein tune the absorption spectrum of retinal? Spectral Tuning in Archaeal Rhodopsins Sensory Rhodopsin II (sRII) Repellent response to blue-green light Spectral features • Absorption maximum is strongly blue-shifted (70 nm from bR). • Prominent sub-band. sRII hR bR 500nm sRI 600nm Sequences Structures of bR and sRII X-ray Structures of bR and sRII Landau et al. Unique opportunity to study spectral shift given by the availability of X-ray structures. • Structures are homologous. (e.g., all-trans retinals) orange: sRII (Natronobacterium pharanois) • Spectra are significantly purple: bR (Halobacterium salinarum) different. Binding Sites of bR and sRII bR Similar structure • Aromatic residues. • Hydrogen-bond network. (counter-ion asparatates, internal water molecules) sRII Mutagenic substitutions (Shimono et al.) T204A/V108M/G130S of sRII produces only 20 nm (30%) spectral shift. What is the main determinant(s) of spectral tuning? Calculation of Absorption Spectra of bR and sRII Combined quantum mechanical/molecular mechanical (QM/MM) calculations. • Retinal is described by ab initio MO (HF/CASSCF). • Protein environment by molecular mechanics force field (AMBER94). Mechanism of Spectral Tuning • Electrostatic interaction between the retinal Schiff base and protein S2 positive charge Me Me S0 S1 S2 Me + + N Me Me H O S1 S1 S0 O C S2 Asp (Glu) S0 isolated in protein • Electronic reorganization of retinal due to polarization of retinal’s wave function Results sRII bR 500nm 600nm S1-S0 : 6.1 (exp. 7.2) kcal/mol. (shift of main absorption band) The shift is mainly due to electronic reorganization. S2-S1 : 1.7 (exp. 4.0) kcal/mol. (appearance of side band in sRII) Optically forbidden in bR, but a peak (side-band) appears in sRII due to intensity borrowing from the S1 state, which is optically allowed. Contributions from Residues Structural Determinants of Spectral Shift G helix is displaced in sRII. Distance between the Schiff base and the counter-ion is shorter. G helix N16 – Cg (Asp201: sRII) : 4.5 A N16 – Cg (Asp212: bR) : 5.2 A QM/MM optimized structures orange: sRII, purple: bR Rhodopsin Photodynamics Quantum (Wave Packets) Dynamic in protein, 1-dimensional surface Ben-Nun et al., Faraday Discussion, 110, 447 - 462 (1998) Calculation of transition amplitude Control of Branching Ratios by Intersection Topography On-the-fly ab initio QM/MM MD Simulation • An analogue of retinal (three double bonds) in bR (20 QM atoms, 96 basis functions) • CASSCF (6,6) / AMBER The Role of Conical Intersection Topography on the Photoisomerization of Retinal Michal Ben-nun Emad Tajkhorshid Shigehito Hayashi Jerome Baudry $$: Beckman Institute, NSF, HFSP, NIH-NCRR