Restriction Mapping of Plasmid DNA
advertisement

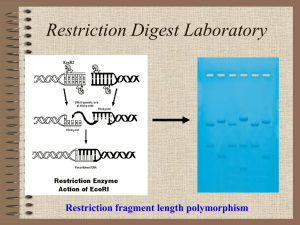
Restriction Mapping of Plasmid DNA Restriction Maps Restriction enzymes can be used to construct maps of plasmid DNA Allows identification of restriction sites found in the sequence of the plasmid Alternative means to discover DNA sequence without sequencing Can be used to determine which restriction enzymes to use for recombinant techniques Getting the Information Isolate the DNA Digest it with restriction enzymes Sort fragments by size using gel electrophoresis Breaks DNA into various size fragments Can sort DNA fragments differing by only a single nucleotide in length Fragment lengths can be used to assemble map of relative positions Digestion Can do single or multiple digestions Single digestion consists of only one enzyme in the mixture Multiple digestions include many or all the enzymes desired Step-wise process gives the most information Digest with individual enzymes and include a multiple digestion Use different sizes of the fragments to construct the map Making the Map The gel will be stained and photographed after running Distances migrated by DNA fragments will be measured from the photograph of the gel A standard curve will be produced by plotting distances migrated by ladder fragments against the size of the fragments Semilog graph paper must be used to obtain a linear plot The standard curve will be use to determine fragment sizes of the digested plasmid DNA A circular map will be constructed from the data