Viral Immunology 2012
advertisement

2/01/12
VIRUS STRUCTURE
Sergei Nekhai, Ph.D.
Objectives:
•Functional organization of viral particles
• Viral Symmetry
• Viral Capsids
Structure of Viruses
• Size range –
– most <0.2 μm; requires electron microscope
Structure of Viruses
Characteristic size scale is
30-100 nm.
Structures are known at
“atomic resolution” - see
Viper website
(http://viperdb.scripps.edu/
Highly symmetric - think
hard about what this
implies about assembly!
(Baker et al.)
Viruses
Figure 13.1
Organization of Viral Particles
•Contains RNA or DNA
Streptococcus
•Form a protective package
E. coli
•Transmit genetic material
•Entry, multiply and exit
the host
•Redirect cellular
machinery
Yeast Cell
Terminology
• Virion: physical virus particle. Nucleocapsid alone for some viruses
(picornaviruses) or including outer envelope structure for others
(retroviruses).
• Capsid (syn: coat): regular, shell-like structure composed of
aggregated protein subunits which surrounds the viral nucleic acid ]
• Nucleocapsid (syn: core): viral nucleic acid enclosed by a capsid
protein coat
• Envelope (syn: viral membrane): lipid bylayer containing viral
glycoproteins. The phospholipids in the bylayer are derived from the
cell that the virus arose from. Not all viruses have envelopes some
consist of only the nucleocapsid
Viruses - Structure
• contain DNA or
RNA
• contain a protein
coat (capsid)
• Some are enclosed
by an envelope
• Some viruses have
spikes
General Structure of Viruses
• Capsids
– All viruses have capsids - protein coats that enclose
and protect their nucleic acid.
– Each capsid is constructed from identical subunits
called capsomers made of protein.
– The capsid together with the nucleic acid are
nucleoscapsid.
The Viral Capsid
• Capsid- Protein coat that encapsidates the viral genome.
• Nucleocapsid-Capsid with genome inside (plus anything
else that may be inside like enzymes and other viral proteins
for some viruses).
Capsid functions
1. Protect genome from atmosphere (May include damaging
UV-light, shearing forces, nucleases either leaked or
secreted by cells).
2. Virus-attachment protein- interacts with cellular receptor to
initiate infection.
3. Delivery of genome in infectious form. May simply “dump”
genome into cytoplasm (most +ssRNA viruses) or serve as
the core for replication (retroviruses and rotaviruses).
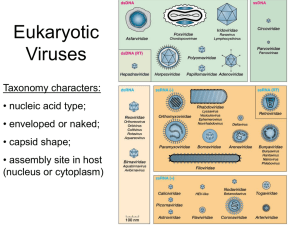
Human Viruses
"Group"
dsDNA
Family
Genome
Genome size (kb) Capsid
Envelope
Poxviridae
Herpesviridae
Adenoviridae
Polyomaviridae
Papillomaviridae
dsDNA, linear
dsDNA, linear
dsDNA, linear
dsDNA, circular
dsDNA, circular
130 to 375
125 to 240
26 to 45
5
7 to 8
Ovoid
Icosahedral
Icosahedral
Icosahedral
Icosahedral
Yes
Yes
No
No
No
Anellovirus
Parvoviradae
ssDNA circular
ssDNA, linear, (- or +/-)
3 to 4
5
Isometric
Icosahedral
No
No
Hepadnaviridae
Retroviridae
dsDNA (partial), circular
ssRNA (+), diploid
3 to 4
7 to 13
Icosahedral
Spherical, rod or cone shaped
Yes
Yes
Reoviridae
dsRNA, segmented
19 to 32
Icosahedral
No
Rhabdoviridae
Filoviridae
Paramyxoviridae
Orthomyxoviridae
Bunyaviridae
Arenaviridae
Deltavirus
ssRNA (-)
ssRNA (-)
ssRNA (-)
ssRNA (-), segmented
ssRNA (-, ambi), segmented
ssRNA (-, ambi), segmented
ssRNA (-) circular
11 to 15
19
10 to 15
10 to 13.6
11 to 19
11
2
Helical
Helical
Helical
Helical
Helical
Circular, nucleosomal
Spherical
Yes
Yes
Yes
Yes
Yes
Yes
Yes
Picornaviridae
Calciviridae
Hepevirus
Astroviridae
Coronaviridae
Flaviviridae
Togaviridae
ssRNA (+)
ssRNA (+)
ssRNA (+)
ssRNA (+)
ssRNA (+)
ssRNA (+)
ssRNA (+)
7 to 9
7 to 8
7
6 to 7
28 to 31
10 to 12
11 to 12
Icosahedral
Icosahedral
Icosahedral
Isometric
Helical
Spherical
Icosahedral
No
No
No
No
Yes
Yes
Yes
ssDNA
Retro
dsRNA
ssRNA (-)
ssRNA (+)
Principles of Viral Architecture
•Viral capsid are made of repated protein subunits
•Capsids are self assembled
•Fraenkel-Conrat and Williams (1955): self-assembly of TMV
•Proteins and nucleic acids are held together with noncovalent bonds
•Protein-protein, protein-nucleic acid, protein-lipid
•Helical or icosahedral symmetry
Viral Capsids
• If 1 protein for 1 capsid:
– Need > 18,000 amino acids.
– Need > 54,000 nucleotides.
– Small viruses hold max. of 5,000 nucleotides.
• Must use many copies of 1 (or a few) protein(s).
• High symmetry
– Minimizes # different subunit interactions involved
with assembly.
– Simpler protein.
– Self assembly:
• Self-contained assembly "instructions".
Basic Nucleocapsid Structures:
• HELICAL: Rod shaped, varying widths and specific
architectures; no theoretical limit to the amount of nucleic
acid that can be packaged
• CUBIC (Icosahedral): Spherical, amount of nucleic acid
that can be packaged is limited by the number of
capsomers and the size of the viral particle
• Irregular: Without clear symmetry
Capsid and Envelope
Non-enveloped
Helical
Icosahedral
Capsid:
•Protect viral nucleic acid
•Interact with the nucleic acid for
packaging
•Interact with vector for specific
transmission
•Interact with host receptors for entry
to cell and to release of nucleic acid
Enveloped
Envelope:
•Made from host cell
membrane (plasma,
ER or Golgi)
•Fuse for Entry
Helical viruses
• Organized around a single axis (the “helix axis”)
• Probably evolved along with other helical structures like
DNA, a-helix, etc.
• Allow flexibility (bending)
• Helical viruses form a closely related spring like helix
instead. The best studied TMV but many animal viruses and
phage use this general arrangement.
– Note-all animal viruses that are helical are enveloped, unlike many of
the phage and plant viruses.
• Most helixes are formed by a single major protein arranged
with a constant relationship to each other (amplitude and
pitch).
• They can be described by their Pitch (P, in nm):
• P= u x p, u-# of protein subunits per helical turn, p-axial rise
per subunit
Helical symmetry
• Tobacco mosaic virus is typical,
well-studied example
• Each particle contains only a single
molecule of RNA (6395 nucleotide
residues) and 2130 copies of the
coat protein subunit (158 amino
acid residues; 17.3 kilodaltons)
– u=16.33 subunits/turn
– p=1.4 Å
– P= 23 Å
• TMV protein subunits + nucleic
acid will self-assemble in vitro in an
energy-independent fashion
• Self-assembly also occurs in the
absence of RNA
TMV rod is 18 nanometers
(nm) X 300 nm
Influenza virus
Ebola Virus
• Filamentous Filovirus with single-stranded (-) RNA genome
• The capsid has a helical morphology and is encased inside a membrane
envelope.
• VP30- matrix protein; L protein – RNA polymerase
Vesicular Stomatitis Virus
• VSV coat protein (50 aa): alpha helical with 3 distinct domains:
+ charge interacts with nucleic acid, hydrophobic with proteins on
either side, negative charge with polar environment
• Subunits are tilted 20o relative to the long axis of the particle.
• VSV Genome: 11,000 nt -ssRNA interacts with the nucleocapsid
protein (N) to form a helical structure with P=5 nm. .
•
ICOSAHEDRAL VIRUSES
•1956, Watson and Crick – only cubic symmetry
leads to isometric particle
•Only three cubic symmetry exist:
•tetrahedral (2:3)
–
12 identical
subunits
•octahedral (4:3:2)
–
24 identical
subunits
•icosahedral (5:3:2)
–
60 identical
subunits
•For viruses of 150-200 Å - ~ 60 of
20 kDa protein subunits
•However, for viruses > 250 Å (turnip
yellow mosaic), it was more than 60
subunits
Parvovirus Structure
Picornavirus Structure
QUASI-EQUIVALENCE
1962, Caspar and Klug – found a principal of building
icosahedral structures from similar blocks
• Shell is built from the same blocks
•Bonds are deformed in a slightly different ways
•Assumed a possibility of 5 degrees deformation
•Shell can contain 60n subunits
A Fuller geodesic dome
That inspired Caspar
and Klug
Triangulation number (T) Enumerated by Caspar
and Klug
• T=f2 x P where f=# of subdivisions on each side of a triangular
face, P=h2 + hk + k2 where h and k are any nonnegative integer
• Only T’s that may be derived from the above equation are
possible.
• 60 = minimal number of irregular subunits required
CLASSES OF ICOSAHEDRAL DELTAHEDRA
Tabulation of the Triangulation Number T
Class
P=1 1
P=3
4
9
3
Skew Classes
16
25
12
7
13
....
27
....
19 21 . . . .
T = Pf2, where P = h2 + hk + k2, h and k any pair of integers with no common
factor, and f= 1 , 2 , 3 , 4 , . . . .
Number of structure units S=60 T
Number morphological units M = 10 T + 2= 10(T-1) hexamers + 12 pentamers
CLASSES OF ICOSAHEDRAL DELTAHEDRA
(a) P=1, T=1. (b) P = 1, T= 4. (c) and (d), P = 3 (T=3 and 12, respectively). (e), (f),
(g) and (h), first members of the skew classes P = 7, 13, 19, and 21, respectively.
Different Arrangements of Icosahedral Symmetry
Zlotnick A. PNAS 2004;101:15549-15550
©2004 by National Academy of Sciences
Jellyroll: Many, but not all Viral
proteins
Capsid proteins
b-barrel.
• Rhombohedral wedges:
– Fit into icosahedron.
• Jellyroll topology
• Conserved in many small
viruses
– T = 1, 3, …
• 60, 180, 240 proteins…
– RNA or DNA viruses.
• Essentially no sequence
homology.
Picornaviridae, a prototype T=3 virus
• Quasi-equivalence with pentamer at each vertex and
hexamers in other regions;
• Triangulation # = 3.
• Note that VP-4 is not on the surface of the structure but
lies under the face.
Picornaviridae, a prototype T=3 virus
• The protein subunits that form each protomer all assume a similar
(not identical) shape .
• In fact all T=3 RNA viruses have proteins that form “8 strand
antiparallel b barrels”.
• The structures form from the polypeptide by first forming a “jelly-roll
barrel” that then goes on to form the wedge-shaped barrel when the
capsid is being formed.
Tomato Bushy Stunt Virus
TBSV icosahedron is 35.4
nm in diameter
• Each particle contains
only a single molecule
of RNA (4800 nt) and
180 copies of the coat
protein subunit (387 aa;
41 kd)
T= 3 Lattice
C
• Viruses similar to TBSV
will self-assemble in
vitro from protein
subunits + nucleic acid
N
in an energyProtein Subunits Capsomeres
independent fashion
Assembly of Turnip Crinkle Virus
Scaffold-guided Assembly of Bacteriophage HK97
T=7
420 subunits
2/03/12 continued- VIRUS
STRUCTURE
Sergei Nekhai, Ph.D.
Objectives:
•Cont-structure of viral capsids
•Enveloped viruses
•Packaging of viral RNA or DNA
•Complex viruses
•Virus maturation, assembly and release
Adenovirus Assembly with Pentons, Hexons and Cement Proteins
T=25
420 subunits
Bluetongue virus
Multishelled Structure of Rotavirus
T=13
Inner shell – VP2, 120 copies
Outer shell – VP6, 780 copies
Reoviridae
• Reoviruses have non-enveloped, icosahedral T = 13 capsids
composed of double protein shell with a complex structure.
• The structure of the bluetongue virus core was recently
reported & represents the largest structure yet determined to
atomic resolution (3.5 Å).
• The outer shell of this virus is approximately 80 nm in
diameter & the inner shell (core) about 60 nm.
• The double-stranded RNA genome of the virus is packed
tightly inside the core surrounding transcription complexes
at the apices of the particle. These genome segments
maintain their order during transcription.
Viral Genome Packaging
• The primary function of the virus particle is to contain
& protect the genome before delivering it to the
appropriate host cell.
• Therefore, the proteins of the capsid must interact
with the nucleic acid genome.
• In most cases, the linear virus genome when stretched
out in solution is at least an order of magnitude longer
than the diameter of the capsid.
• Merely folding the genome in order to stuff it into
such a confined space is complex, but is compounded
by the fact that repulsion by the cumulative negative
electrostatic charges on the phosphate groups of the
nucleotide backbone mean that the genome resists
being crammed into a small space.
Viral Genome Packaging
• Viruses package, along with the genome, a number of
positively charged molecules to counteract negative charges.
• These include small, positively charged ions (Na, Mg, K, etc.),
polyamines & various nucleic acid-binding proteins.
• Some of these proteins are virus-encoded & contain amino
acids with basic side-chains such as arginine & lysine which
interact with the genome, e.g. retrovirus NC & rhabdovirus N
(nucleocapsid) proteins, & influenza virus NP protein
(nucleoprotein).
• Many viruses with double-stranded DNA genomes have basic
histone-like molecules closely associated with the DNA.
Some are virus-encoded, e.g. adenovirus polypeptide VII.
Other viruses utilize cellular proteins, e.g. the polyomavirus
genome assumes a chromatin-like structure in association with
four cellular histone proteins, H2A, H2B, H3 & H4, similar to
that of the host cell genome.
Viral Genome Packaging
• Another problem viruses must overcome is how to achieve
the specificity required to select & encapsidate the virus
genome from the large background of cellular nucleic acids.
• In most cases, by the late stages of virus infection when
assembly of virus particles occurs, transcription of cellular
genes has been reduced & a large pool of virus genomes
have accumulated - overproduction of virus nucleic acids
eases but does not eliminate the problem of specific genome
packaging.
• Therefore, a specific virus-encoded capsid or nucleocapsid
protein is required to achieve this end & many viruses, even
those with relatively short, compact genomes such as
retroviruses & rhabdoviruses, encode this type of protein.
Viral Genome Packaging
• During particle assembly, viruses frequently make
mistakes.
• These can be physically measured by
particle:infectivity ratios, i.e. the ratio of the total
number of particles in a virus preparation (counted by
electron microscopy) to the number of particles able to
give rise to infectious progeny (measured by plaque or
limiting dilution assays).
• This value is in some cases found to be several
thousand particles to each infectious virion & only
rarely approaches a ratio of 1:1.
Viral Genome Packaging
• Specific nucleotide sequences in the genome (the packaging
signal) permit the virus to select genomic nucleic acids from
the cellular background.
• The packaging signal from a number of virus genomes has
been identified, e.g. the Y ('psi') signal in murine retrovirus
genomes & the sequences responsible for packaging the
genomes of several DNA virus genomes (some adenoviruses
& herpesviruses) have been clearly & unambiguously
defined.
• Accurate & efficient genome packaging requires information
also from regions of secondary structure formed by the
folding of the genomic nucleic acid into complex forms.
Positive-strand RNA Genome Packaging
• Simple ssRNA genomes (plant viruses,
picornaviruses, alphaviruses) – no specific overall
fold, pack tightly highly structured RNA
• May pack into shallow groove of the capsid
• Recognition – MS2, a dimer of a coat protein binds a
hairpin
• None-enveloped eukaryotic coat proteins extend a
flexible positively charged arm that recognize RNA
(alfalfa virus)
Packaging of MS2 virus (T3)
Packaging of Alfalfa Mosaic Virus
Packaging of HIV-1
(c)
(a) Amino acid sequence of the HIV-1pNL4–3 nucleocapsid protein showing the zinc binding mode of the
CCHC zinc knuckles. (b) Nucleotide sequence and secondary structure of the HIV-1pNL4–3 Ψ-site. The
sequence of the SL2 RNA construct used here is shown in bold letters, and residues of the major splice donor
site are denoted by open letters. (c) A representative NC-SL2 structure. The nucleobases of residues G9, U10
and G11 are colored green, orange and purple, respectively. The side-chains of selected basic residues are
colored blue, and the zinc atoms are displayed as silver spheres. The N-terminal zinc knuckle packs against
A15 of the A5-U14-A15 base triple, and the side-chains of Lys34 and Lys47 are poised to form salt-bridges
with phosphodiester groups.
Amarasinghe et al., 2000 J Mol Biol, 301: 491-511
Genome Packaging
• Rather less is known about the arrangement of the genome inside
virus particles with icosahedral symmetry.
• T = 3 icosahedral RNA virus capsid subunits consist largely of
the '8-strand anti-parallel b-barrel' structural motif, discussed
earlier.
• In these viruses, positively-charged inward-projecting arms of
the capsid proteins interact with the RNA in the center of the
particle.
• BPMV (bean pod mottle virus), a T = 3 Comovirus with a
bipartite genome. X-ray crystallography showed that the RNA is
folded in such a way that it assumes icosahedral symmetry,
corresponding to that of the capsid surrounding it.
• The regions which contact the capsid proteins are single-stranded
& appear to interact by electrostatic forces rather than covalent
bonds. The atomic structure of X74 also shows that a portion of
the DNA genome interacts with arginine residues exposed on the
inner surface of the capsid in a manner similar to BPMV.
Genome Packaging in TMV
• TMV, (+)sense RNA helical plant virus
• Only has a single major coat protein, & will
spontaneously assemble from its purified RNA & protein
components in vitro.
• Particle assembly is initiated by association of preformed
aggregates of coat protein molecules ('discs') with
residues 5444-5518 in the 6.4 kb RNA genome, known as
the origin of assembly sequence (OAS).
• The flat discs have 17 subunits per ring, close to the 16.34
subunits per turn found in the mature virus particle.
Genome Packaging in TMV
Genome Packaging in TMV
• The discs have a pronounced polarity. Assembly begins when a
disc interacts with the OAS in genomic RNA.
• This converts the discs to a helical 'locked washer' structure, each
of which contains 3' coat protein subunits.
• Further discs add to this structure, switching to the 'locked
washer' conformation.
• RNA is drawn into the assembling structure in what is known as a
'travelling loop', which gives the common name to this
mechanism of particle formation - the vRNA is trapped &
subsequently buried in the middle of the disc as the helix grows.
• Extension of the helical structure occurs in both directions but at
unequal rates. Growth in the 5' direction is rapid because a disc
can add straight to the protein filament & the travelling loop of
RNA is drawn up through it. Growth in the 3' direction is slower
because the RNA has to be threaded through the disc before it can
add to the structure.
dsDNA Genome Packaging
• dsDNA packaging of tailed bacteriophages – DNA
insertion into icosahedrical head is ATP dependent,
driven by a motor
• Herpesvirus – rolling circle replication
• Adenovirus – AT-rich repeat that determines
incorporation into
• PRD1 (similar to adenovirus) – unique vertex
containing a protein needed for DNA packaging, that
also includes ATPase
• Papovaviruses – incorporate cellular histones, to form
20-25 nucleosomes
Genome Packaging in M13
• Enterobacteria phage M13 is
another helical virus where
protein-nucleic acid
interactions in the virus
particle are relatively simple.
• The primary sequence of the
g8p molecule determines the
orientation of the protein in
the capsid.
• The inner surface of the rodlike phage capsid is positively
charged & interacts with the
negatively charged genome,
while the outer surface of the
cylindrical capsid is
negatively charged.
Genome Packaging in M13
• During replication, the genomic DNA is associated with a nonstructural DNA-binding protein, g5p.
• g5p is the most abundant of all virus proteins in the infected E.
coli cell, & coats the newly replicated single-stranded phage DNA,
forming an intracellular rod-like structure similar to the mature
phage particle.
• The function of g5p is to protect the genome from host cell
nucleases & to interrupt genome replication, sequestering newly
formed strands as substrates for encapsidation.
• Newly synthesized coat protein monomers (g8p) are associated
with the inner (cytoplasmic) membrane of the cell & it is at this
site that assembly of the virus particle occurs.
• The g5p coating is stripped off as the particle passes out through
the membrane & is essentially exchanged for the mature g8p coat
(plus the accessory proteins).
• The protein-nucleic acid interactions which occur appear to be
rather simple & involve opposing electrostatic charges & the
stacking of the DNA bases between the planar side-chains of the
proteins.
dsRNA Genome Packaging
• Reovirus – 12 RNA segments, selection signal is
unclear, dsRNA is tightly packaged
• Bacteriophage j5 – sequential incorporation of three
dsRNA segments. Includes ATPas and assembly
clamp
• ATPase could be a unwinding motor to package
ssRNA instead of dsRNA
Negative-strand RNA Genome Packaging
• Influenza – eight-segment genome packaged with the
help of N protein. Form rods, 300- 1,200 angstroms.
Packaging of dsDNA and dsRNA viruses
The Structure of a Herpesvirus
Tegument
Icosahedral cores
Spikes
Envelope
Enveloped viruses
• In an enveloped virus, the capsid is covered by an
envelope
– The envelope is usually made of some combination of lipids,
proteins, and carbohydrates
– Some envelopes contain spikes that allow them to attach to the
host
Enveloped Viruses
• Enveloped Viruses
• Influenzavirus
• herpes simplex
virus
Enveloped Viruses
• Many viruses have devised strategies to exit from
the infected cell without its total destruction.
• All living cells are covered by a membrane
composed of a lipid bilayer - the viability of the
cell depends on the integrity of this membrane.
Viruses leaving the cell must, therefore, allow this
membrane to remain intact.
• This is achieved by extrusion (budding) of the
particle through the membrane, during which
process the particle becomes coated in a lipid
envelope derived from the host cell membrane &
with a similar composition.
Viral
Budding
Formation of enveloped virus particles
• The structure underlying the envelope may be based
on helical or icosahedral symmetry & may be formed
before or as the virus leaves the cell.
• In the majority of cases, enveloped viruses use cellular
membranes as sites allowing them to direct assembly.
• The formation of the particle inside the cell,
maturation & release are in many cases a continuous
process.
• The site of assembly varies for different viruses - not
all use the cell surface membrane; many use
cytoplasmic membranes such as the Golgi apparatus,
others, such as herpesviruses, which replicate in the
nucleus may utilize the nuclear membrane. In these
cases, the virus is usually extruded into some form of
vacuole, in which it is transported to the cell surface &
subsequently released.
Viral Structure: Envelope
Proteins
Envelope proteins
• If the virus particle became covered in a smooth,
unbroken lipid bilayer, this would be its undoing.
• Such a coating is effectively inert, & although
effective in preventing desiccation of or enzymatic
damage to the particle, would not permit recognition
of receptor molecules on the host cell.
• Therefore, viruses modify their lipid envelopes by the
synthesis of several classes of proteins which are
associated in one of three ways with the envelope.
Viral Structure
• Matrix proteins: are internal virion proteins
whose function is effectively to link the internal
nucleocapsid assembly to the envelope. Such
proteins are not usually glycosylated & are often
very abundant, for example, in retroviruses they
comprise approximately 30% of the total weight
of the virion.
• Glycoproteins: are transmembrane proteins
anchored to the membrane by a hydrophobic
domain, & can be subdivided into two types by
their function:
• External glycoproteins are anchored in the envelope by a
single transmembrane domain.
• Transport channel proteins contain multiple hydrophobic
transmembrane domains, forming a protein-lined channel
through the envelope.
Fusion of Bilayer Membrane
Hemifusion
Stalk
Transition
Structure
Hemifusion
diaphragm
Fuision
pore
Fusion of Class I Fusion Proteins
Fusion of Class II Fusion Proteins
Complex Virus Structures
• However, there are many viruses whose structure is
more complex than those with helical symmetry or
icosahedral symmetry.
• In these cases, although the general principles of
symmetry are often used to build part of the virus
shell, the larger & more complex viruses cannot be
simply defined by a mathematical equation as can a
simple helix or icosahedron.
• Because of the complexity of some of these viruses,
they have defied attempts to determine detailed atomic
structures.
Poxvirus Particle
Poxviruses
• Example of complex viral structure-Poxviridae.
• These viruses have oval or 'brick-shaped' particles
200-400 nm long.
• These particles are so large that they were first
observed in using high-resolution optical microscopes
in 1886, & thought at that time to be 'the spores of
micrococci'.
• The external surface of the virion is ridged in parallel
rows, sometimes arranged helically.
• The particles are extremely complex & have been
shown to contain more than 100 different proteins.
Poxviruses
• Under the electron microscope, thin sections of poxviruses
reveal that the outer surface of the virion is composed of lipid &
protein.
• This surrounds the core, which is biconcave (dumbbell-shaped),
& two 'lateral bodies' whose function is unknown.
• The core is composed of a tightly compressed nucleoprotein &
the double-stranded DNA genome is wound around it.
• Antigenically, poxviruses are very complex, inducing both
specific & cross-reacting antibodies - hence the possibility of
vaccinating against one disease with another virus (e.g. the use
of Vaccinia virus to immunize against smallpox (variola) virus).
• Poxviruses & a number of other complex viruses also emphasize
the true complexity of some viruses - there are at least 10
enzymes present in poxvirus particles, mostly involved in
nucleic acid metabolism/genome replication.
Poxviruses
•During replication, two forms of poxvirus particle are
observed:
–extracellular forms which contain two membranes
–intracellular particles which only have an inner membrane
•Poxviruses & other virus with complex structures obtain
their membranes in a different way from "simple" enveloped
viruses such as retroviruses or influenza.
•Rather than budding at the cell surface or into an
intracellular compartment, acquiring a single membrane,
these complex viruses are wrapped by the endoplasmic
reticulum, acquiring two layers of membrane.
Bacteriophages
• Complex virus
– Head is polyhedral
– Tail is helical
– It is surrounded by a
protein coat (capsid)
Figure 13.5a
Caudovirales:
Myoviridae, Siphoviridae & Podoviridae
• The tailed phages of enterobacteria have been extensively
studied for excellent reasons - easy to propagate in bacterial
cells, can be obtained in high titres, & are easily purified,
facilitating biochemical & structural studies.
• The head of the particles consists of an icosahedral shell with T
= 7 symmetry, attached by a collar to a contractile, helical tail.
At the end of the tail is a plate which functions in attachment to
the bacterial host & also in penetration of the bacterial cell wall
using lysozyme-like enzymes associated with the plate.
• Thin protein fibres attached to the plate & the tail plate itself are
involved in binding to the receptor molecules in the wall of the
host cell.
The Caudovirales
• There are separate
assembly pathways
for the head & tail
sections of the
particle, which
come together at a
late stage to make
up the virion.
• These viruses
illustrate how
complex particles
can be built up from
the simple
principles outlined
before.
Geminiviridae
• Another example is
provided by the structure
of geminivirus particles,
which consist of two
twinned T = 1 icosahedra.
• Each icosahedron has one
morphological subunit
missing & the icosahedra
are joined so the mature
particle contains 110
protein monomers
arranged in 22
morphological subunits.
Baculoviridae
Baculoviridae
• Baculoviruses contain 12-30 structural proteins, &
consist of a rod-like (hence “baculo”) nucleocapsid 3060 nm diameter & 250-300 nm long which contains the
88-160 kbp double-stranded DNA genome.
• The nucleocapsid is surrounded by an envelope, outside
which there may or may not be a crystalline protein
matrix. If this outer protein shell is present, the whole
assemblage is referred to as an 'occlusion body' & the
virus is said to be occluded.
• The function of the occlusion bodies is to confer
resistance to adverse environmental conditions, which
enables the virus to persist in soil or on plant materials
for extended periods of time waiting to be ingested by a
new host.
Structural Heterogeneity of HIV
Particles
Mean diameter approximately
120nm.
Average volume approximately
10^6 nm^3
Approximate mass per virus
particle is 1fg (650 Mda)
Note that the structures of these
viruses are a lot more sloppy
than the structures of the
icosahedral viruses we
discussed earlier.
Red corresponds to conical,
orange to rod shapes and
yellow is psycho.
Each picture is 160nm wide.
Benjamin et al.
A Reminder on Cryo Electron
Microscopy
By rotating a sample through a number of different orientations it is possible
to generate a series of images which with careful analysis can be used to
garner a three-dimensional picture of macromolecular complexes
(ribosomes, viruses, etc.)
HIV-1 Capsid Structure
Fascinating structure of the internal
capsid of HIV.
Shape conferred by geometric
rules about 5-fold defects.
Sundquist et al.
HIV-1 Capsid
Assembly
• Assembly involves the collection of all the components necessary
for the formation of the mature virion at a particular site in the
cell.
• During assembly, the basic structure of the virus particle is
formed.
• The site of assembly depends on the site of replication within the
cell & on the mechanism by which the virus is eventually released
from the cell & varies for different viruses:
• in picornaviruses, poxviruses & reoviruses assembly occurs in the
cytoplasm
• in adenoviruses, polyomaviruses & parvoviruses it occurs in the
nucleus
• As with the early stages of replication, it is not always possible to
identify the assembly, maturation & release of virus particles as
distinct & separate phases.
Maturation
• Maturation is the stage of the replication-cycle at which the
virus becomes infectious.
• Maturation usually involves structural changes in the virus
particle which may result from specific cleavages of capsid
proteins to form the mature products or conformational
changes in proteins during assembly.
• Such events frequently lead to substantial structural
changes in the capsid which may be detectable by criteria
such as differences in the antigenicity of incomplete &
mature virus particles or the condensation of nucleoproteins
with the virus genome.
• Virus proteases are frequently involved in maturation,
although cellular enzymes or a mixture of virus & cellular
enzymes are used in some cases.
Release
• Apart from plant viruses which have evolved particular
strategies to overcome the structure of plant cell walls, all
other viruses escape the cell by one of two mechanisms:
• For lytic viruses (most non-enveloped viruses), release is
a simple process - the infected cell breaks open & releases
the virus.
• Enveloped viruses acquire their lipid membrane as the
virus buds out of the cell through the cell membrane or
into an intracellular vesicle prior to subsequent release.
Virion envelope proteins are picked up during this
process as the virus particle is extruded - this process is
known as budding.
Maturation: Naked Icosahedral Viruses
•Preassembled capsomers are joined to
form empty capsids (procapsid)
•The assembly of capsomers to form
the procapsid is often accompanied by
extensive reorganization, which is
revealed by changes in serological
specificity and isoelectric point. eg.
picornaviruses and adenoviruses.
Release: Naked Icosahedral Viruses
Poliovirus - released, with lysis of infected cells
Virions of DNA viruses - mature in the nucleus
and tend to accumulate within infected cells over
a long period and are released when the cell
undergoes autolysis, and in some cases, may be
extruded without lysis.
Maturation: Enveloped Viruses
•Viral proteins are first associated with the nucleic
acid to form nucleocapsid
•Nucleocapsid is then surrounded by an envelope
• In nucleocapsid formation, the proteins are all
synthesized on cytoplasmic polysomes and are
rapidly assembled into capsid components
• In envelope assembly, virus-specified envelope
proteins go directly to the appropriate cell membrane
(the plasma membrane, the ER, the Golgi apparatus),
displacing host proteins.
Maturation: Enveloped Viruses
•Viral envelope is made of carbohydrates and lipids produced by
the host cell (eg. the plasma membrane for orthomyxoviruses and
paramyxoviruses, the nuclear membrane for herpesviruses)
•A given virus will differ in its lipids and carbohydrates when
grown in different cells, with consequent differences in physical,
biological, and antigenic properties
•The envelope glycoproteins are synthesized in the following
manner: the polypeptide backbone is first formed on polysomes
bound to the ER, which then moves via transport vesicles to the
Golgi apparatus where it attains it full glycosylation and fatty acid
acylation.
•The matrix proteins that are present in viral envelope are usually
not glycosylated and stick to the cytoplasmic side of the plasma
membrane through hydrophobic domains
Maturation: HIV-1
Virion Enzyme Activities
• Virions are not inert structures.
• Many virus particles contain one or more enzymatic activities,
although in most cases these are not active outside the
biochemical environment of the host cell.
• All viruses with negative-sense RNA genomes must carry
with them a virus-specific RNA-dependent RNA polymerase
because most eukaryotic cells have no mechanism for RNAdependent RNA polymerization - genome replication could
not occur if this enzyme were not included in the virus
particle.
• Reverse transcription of retrovirus genomes occurs inside a
particulate complex & not free in solution.
• The more complex DNA viruses (e.g. herpesviruses &
poxviruses) carry a multiplicity of enzymes, mostly concerned
with some aspect of nucleic acid metabolism.
HIV-1 Restriction Factors: APOBEC3G and TRIM5a
Questions:
snekhai@howard.edu
Lecture Slides:
www.sicklecell.howard.edu/research.htm