PBL2: Restriction Mapping
advertisement

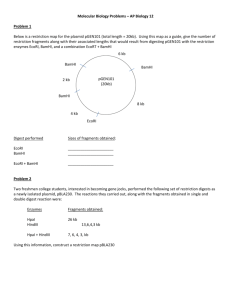
PBL2: Restriction Mapping 1. Define restriction endonuclease. 2. Complete the following table Site Structure pUC19 TaqI T^CGA 4 " DdeI C^TNAG 6 " PvuII CAG^CTG 2 " HaeII RGCGC^Y 3 pBR322 BamHI G^GATCC 1 " 3. # expected # expected # of sites (equal base (known base observed content) content) Plasmid RsaI GT^AC 3 Plasmid pBR322 was first digested with EcoRI, which has a unique site within the plasmid. The plasmid was then treated with two different restriction enzymes (A & B), both as single and double digests. The sizes of the fragments obtained by each digest were determined by gel electrophoresis as follows: Enzyme A Enzyme B Both Enzymes 650 bp 240 bp 240 bp 1590 880 280 2120 930 650 2310 880 1070 1240 a. b. c. 4. Construct a restriction map of pBR322 showing the positions of the restriction sites for EcoRI and these two enzymes. Is this a unique map? Suggest the identities of enzymes A and B. Plasmid pBR322 was first digested with EcoRI. The plasmid was then partially digested with restriction enzyme X. From the partial digest, fragments of the following sizes were found: 190, 350, 540, 680, 880, 1080, 1220, 1760, 2600, 3280, 3470, 3820 & 4360 base pairs. a. Construct a restriction map of pBR322 showing the positions of the restriction sites for EcoRI and enzyme X. b. Is this a unique map? c. Suggest the identity of enzyme X. 5. What systematic process did you use to construct the restriction maps? Is this process an algorithm? If not, why not? If yes, is it of type NP?