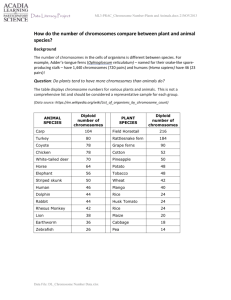
Cytogenetics: Nomenclature and Disease
advertisement

Cytogenetics: Nomenclature and Disease Willis Navarro, MD Medical Director, Transplant Services National Marrow Donor Program Overview • Normal Chromosomes – Structure – Genes • Chromosomal Disruptions – Types off Chromosomal C Changes C • Disruptions and Disease Structural Overview • DNA forms a double helix • Double helix structure is wound around histones • DNA/histone complex th fforms the then th chromosome structure http://ghr.nlm.nih.gov/handbook/illustrations/chromosomestructure Cell Division and Cytogenetics • Tissue cells of interest are grown in culture • Cell must be “frozen” at metaphase t h – Mitotic inhibitor added – Chromosomes condensed – Cells harvested From: http://www.google.com/imgres?imgurl=http://nte-serveur.univ-lyon1.fr/nte/EMBRYON/www.uoguelph.ca/zoology/devobio/miller/mitosis1.gif&imgrefurl=http://nte-serveur.univlyon1.fr/nte/EMBRYON/www.uoguelph.ca/zoology/devobio/210labs/mitosis1.html&h=538&w=450&sz=24&tbnid=PXVEm8paeVwJ::&tbnh=132&tbnw=110&prev=/images%3Fq%3Dmitosis&hl =en&usg=__XTdVA_DgEhY6qk-d9HDMvHcVN9Q=&sa=X&oi=image_result&resnum=4&ct=image&cd=1 Human Chromosome Basics C metacentric submetacentric C C Acrocentric From: http://www.miscarriage.com.au/images/pages/karyotype_normal.jpg • • • 22 pairs plus 2 sex chromosomes (diploid number: 46): (46, XX) Composed of DNA plus infrastructure (histones, proteins, RNA, sugars) 3 groups of shapes based on centromere position, arm length Example of G-Banding: Ch 11 Chromosome p arm Centromere q arm • GTL stain: Giemsa/Trypsin/Leishman • Chr 11 is submetacentric • Representative R t ti ideogram id • Stained to distinguish denser and less dense areas • Unique staining patterns for each chromosome • Many genes coded • Banding ≠ genes From: http://upload.wikimedia.org/wikipedia/commons/thumb/c/cf/Chromosome_11.svg/164px-Chromosome_11.svg.png How Do You Define a Gene? • DNA sequence q begins with a start codon; ends with a stop p codon • Amino acids (each with a 3-character code) then join to form a protein which then has a function • There Th is i “filler” “fill ” DNA that codes for other stuff So Many Genes Genes… • p arm Centromere q arm Dessen P, Knuutila S, Huret JL Chromosome 11. Atlas Genet Cytogenet Oncol Haematol. 2002. URL : http://AtlasGeneticsOncology.org/Indexbychrom/idxa_11.html © Atlas of Genetics and Cytogenetics in Oncology and Haematology ARHGAP20 109952.97611q23.2ARHGEF12 119713.15611q23.3ATM 107598.76911q22-q23BAD 63793.87811q13.1BIRC3 101693.40411q22C11orf30 75833.71711q13.5CARS 2978.73511p15.5CASP1 104401.44711q23CBL 118582.20011q23.3-qterCCND1 69165.05411q13CHKA 67576.90211q13.1DDB2 47193.06911p12-p11DDX10 108041.02611q22-q23EXT2 44073.67511p12-p11FANCF 22600.65511p15HRAS 522.24211p15.5KAT5 65236.06511q13LMO2 33836.69911p13MAML2 95351.08811q21MEN1 64327 57011q13MIRN125B1 121475.67511MLL 64327.57011q13MIRN125B1 121475 67511MLL 117812.41511q23MRE11A 117812 41511q23MRE11A 93790.11511q21MYEOV 93790 11511q21MYEOV 68818.19811q13.2NUMA1 71391.55911q13NUP98 3689.63511p15PAK1 76710.70811q13-q14PICALM 85346.13311q14POU2AF1 110728.19011q23.1RELA 65178.39311q13RSF1 77054.92211q13.5SDHD 111462.83211q23SPA17 124048.95011q24.2SPI1 47332.98511p12-p11.22WT1 32365.90111p13ZBTB16 113436.49811q23 ACAT1 107497.46811q22.3ACP2 47217.42911p11.2ADAMTS15 129824.07911q25ADAMTS8 129780.02811q25ADM 10283.21811ADRBK1 66790.48111q13AHNAK 62039.95011q12-q13AIP 67007.09711q13.3ALDH3B2 67186.20911q13ALKBH3 43858.97311p11.2ALKBH8 106880.54411q22.3AMPD3 10428.80011p15ANKK1 112763.72311q23.2ANO1 69602.05611q13.2APBB1 6372.93111p15API5 43290.10911APLNR 56757.64311q12APOA1 116211.67911q23-q24ARAP1 72073.76211q13.3ARCN1 117948.32111q23.3ARFIP2 6453.50211p15ARHGAP1 46655.20811p11.2ARHGEF17 46655 20811p11 2ARHGEF17 72697.31711q13.3ARL2 72697 31711q13 3ARL2 64538.16211q13ARRB1 64538 16211q13ARRB1 74654.13011q13ART1 74654 13011q13ART1 3622.93711p15ASCL2 2246.30411p15.5B3GAT1 133753.60811q25B3GNT6 76423.08311q13.4BARX2 128751.09111q24.3BCL9L 118272.06111q23.3BDNF 27633.01811p14.1BIRC2 101723.17611q22BRMS1 65861.38011q13-q13.2BRSK2 1367.70511p15.5BTG4 110843.46611q23C11orf17 8889.27711p15.3C11orf68 65440.85911q13.1C1QTNF4 47567.79211q11C1QTNF5 118714.86211q23.3CADM1 114549.55511q23.2CALCA 14946.62411p15.2CAPN1 64705.91911q13CASP4 104318.80411q22.2q22.3CASP5 104370.17211q22.2-q22.3CAT 34417.05411p13CCDC73 32580.20211p13CCKBR 6237.54211p15.4CD151 822.95211p15.5CD248 65838.53411q13CD3E 117680.50511q23CD44 35116.99311p13CD5 60626.50611q13CD59 33681.13211p13CD6 60495.69111q12.2CD81 2355.12311pterp11.2CD82 44543.71711p11.2CDC42EP2 64838.90711q13CDK2AP2 67030.54411q13CDKN1C 2861 02411p15 5CDON 125331.92311q23 2861.02411p15.5CDON 125331 92311q23-q24CENTD2 q24CENTD2 72073.76211q13.3CEP57 72073 76211q13 3CEP57 95163.29011q21CFL1 95163 29011q21CFL1 65378.86111q13.1CHEK1 125000.24611q24.2CHORDC1 89574.26511q14.3CHRDL2 74085.12211CKAP5 46721.66011p11.2CLCF1 66888.21511q13.3CLP1 57181.20611q12CNTF 58146.72111q12CNTN5 98397.08111q21-q22.2COP1 104417.263-COX8A 63498.65511q12-q13CPT1A 68278.66411q13.2CREB3L1 46255.80411q11CRTAM 122214.46511q24.1CRY2 45825.24511p11.2CRYAB 111284.56011q22.3-q23.1CST6 65536.03811q13CTNND1 57285.81011q12.1CTSD 1730.56111p15.5CTSF 66087.51111q13.2CTTN 69922.26011q13CUL5 107384.61811q22.3CXCR5 118259.75111q23.3CYB5R2 7642.90211p15.4DCPS 125678.85711q24DDB1 60823.49511q12-q13DDX25 125279.48211q24DDX6 118123.68311q23.3DEAF1 634.22511p15.5DGKZ 46339.72111p11.2DKK3 11941.11911p15.3DLAT 111400.74811q23.1DNAJC4 63754.32911q13DPF2 64857.92211q13.1DPP3 66004.45611q12-q13.1DRD2 112785.52711q23DUSP8 102485.37011q21-q22.1EED 85633.46311q14.2-q22.3EHF 1531.85711p15.5DYNC2H1 102485.37011q21 q22.1EED 85633.46311q14.2 q22.3EHF 34599.24411p12EI24 124944.50811q23EIF3F 7965.44311p15.4EIF4G2 10775.16911p15ELF5 34456.91811p13-p12ESRRA 63829.62011q12ETS1 127833.87011q23.3F2 46697.31911p11-q12FADD 69726.91711q13.3FADS2 61352.28911q12.2FAM111B 58631.28611q12.1FAM89B 65096.39611q23FAT3 91724.91011q14.3FAU 64644.67811q13FBXL11 66644.07411q13.1FCHSD2 72225.43811q13.3FEN1 61316.72611q12FEZ1 124820.85811q24.2FGF19 69222.18711q13.1FGF3 69333.91711q13FGF4 69296.97811q13.3FLI1 128069.02311q24.1-q24.3FLRT1 63627.93811q12-q13FOLH1 49124.76311p11.2FOLR1 71578.60711q13.3q14.1FOLR2 71605.46711q13.3-q14.1FOSL1 65416.26811q13FOXR1 118347.62711q23.3FRAG1 3786.36011p15.5FSHB 30209.13911p13FTH1 61488.33311q13FUT4 93916.77511q12-qterFXC1 6459.25311p15.2-q15.5FZD4 86334.36911q14-q21GAB2 77603.99011q14.1GAL 68208.55911q13.2GAS2 22652.93111p14.3GCRG224 82211.05611q13 q14GNG3 62231.70911p11GRM5 87880.62611q14.3GSTP1 82211.05611q13-q14GNG3 67107.64211q13-qterGTF2H1 18300.71911p15.1-p14H19 1972.98311p15.5H2AFX 118469.79511q23.3HBB 5203.27211p15.5HCCA2 1447.26911p15.5HEPACAM 124294.35611q24.2HEPN1 124294.356-HIPK3 33235.74411p13HPS5 18256.79311p14HRASLS2 63076.81811q12.2HRASLS3 63098.52011q12.3HSPA8 122433.41011q24.1HSPB2 111288.70911q22-q23HTATIP2 20341.86511HTR3A 113351.12011q23.1q23.2HYOU1 118420.10611q23.1-q23.3ICEBERG 104513.87911q21-q22IFITM1 303.99111p15.5IFITM3 309.67311p15.5IGF2 2106.92311p15.5IGF2AS 2118.31311p15.5IGHMBP2 68427.89511q13.3IL10RA 117362.31911q23IL18 111519.18611q22.2-q22.3ILK 1132.47411p15.5MUC5B 1200.87211p15.5MUC6 1002.82411p15.5MUS81 65384.44811q13MYOD1 17697.68611p15NAALAD2 89507.46611q14.3-q21NAP1L4 … What can go wrong with a gene? • The correct sequence is critical to coding the right protein/protein structure • If the chromosome carrying a particular gene is altered, then the resulting mutated protein or control elements may cause problems What can go wrong with a chromosome? h ? • Constitutional vs acquired q abnormalities • Numerical abnormalities – Monosomy: loss of a whole chromosome – Trisomy: gain of a whole chromosome • Structural abnormalities – Deletions – Inversions – Translocations Monosomy X: Turner Syndrome C Constitutional i i lL Loss Trisomy 21: Down Syndrome C i i lG i Constitutional Gain Deletion http://ghr.nlm.nih.gov/handbook/illustrations Deletion 5q A Acquired i dL Loss • Interstitial losses of the long arm of chromosome 5 • These Th llosses result l iin large numbers of genes being lost • Often associated with myelodysplastic syndromes and acute myeloid leukemia From: http://atlasgeneticsoncology.org/Educ/Images/GeneticCancerFig7.jpg Inversions http://ghr.nlm.nih.gov/handbook/illustrations Inversion (3)(q24q27) A i d Ab li Acquired Abnormality From: http://members.aol.com/chrominfo/images/inv3ideo.gif • Interstitial I t titi l segmentt inverts i t Translocations http://ghr.nlm.nih.gov/handbook/illustrations Translocation t(9;22) A Acquired i d Ab Abnormality li • Material is exchanged between chromosomes 9 and 22 creating a new 22, fusion gene: bcr/abl • Breakpoint may vary a bit such that the newly created fusion protein may be of several lengths From: http://members.aol.com/chrominfo/images/inv3ideo.gif http://atlasgeneticsoncology.org/Anomalies/CML.html – p190 (kDa) – p210 Dicentric Chromosome http://ghr.nlm.nih.gov/handbook/illustrations Isochromosomes http://ghr.nlm.nih.gov/handbook/illustrations Ring Chromosomes http://ghr.nlm.nih.gov/handbook/illustrations Duplication http://ghr.nlm.nih.gov/handbook/illustrations Recap of Basic Abnormalities • Loss or gain of entire chromosomes – Monosomy – Trisomy • Structural – Deletions – Inversions I i – Translocations • Plus more uncommon types of abnormalities – Derivative chromosome (der) • Used when only one chromosome of a translocation is present or • One chromosome has two or more structural abnormalities – – – – – – – Dicentric chromosome (dic) [chromosome has two centromeres] Duplicate (dup) [duplication of a portion of a chromosome] Insertion (ins) Isochromosomes (i) [both arms are the same] Marker chromosome (mar) [unidentifiable piece of chromosome] Ring chromosome (r) Hyperdiploidy: greater than 48 chromosomes Interpreting Cytogenetic Reporting • In sequence: q – – – – – – the overall number of chromosomes identified sex chromosomes affected chromosomes type of abnormalities described in shorthand chromosomal band location In brackets, the number of cells with a given karyotype • Examples – – – – – 46, XX; t(9;22)(q13;q22) [20] 47 XY; +21 [12] 47, 46, XX; inv 16(q13; q21) [20] 45, XY; -5 [18]; 46, XY [2] 46 XY 46, XY; -5 5 (q13) [4] [4]; 46 46, XX [16] Cytogenetic Pioneers Barbara McClintock – Fi Firstt genetic ti map off maize i – Genetic and physical characteristics correlated – Her work helped explain how cells that share the same genome can have differentt ffunctions h diff ti – Nobel Prize for transposons in 1983 Janet Rowley – Hypothesized that leukemias might contain non-random non random genetic abnormalities – 1972: Showed that recurring chromosomal abnormalities occurred in leukemia and sometimes defined the disease’s characteristics Leukemias and Cytogenetics • • Certain morphologic subtypes were known to have distinct prognoses and/or clinical syndromes (M0-M7) Examples: – acute p promyelocytic y y leukemia ((M3)) • High bleeding risk due to coagulopathy but favorable prognosis • t(15; 17)(q22;q12) – AML, subtype M4eo • Favorable prognosis • inv(16)(p13;q22) – Chronic myeloid leukemia • t(9;22)(q11;q34) ( ; )(q ;q ) – Some myelodysplastic patients— typically older women--had a pattern of normal platelet counts and a favorable prognosis • 5q- syndrome Risk Stratification for Acute L k i U Leukemias Using i C Cytogenetics i • Previous to Janet Rowley and others’ observations about cytogenetics and prognosis, leukemias were only categorized by morphology under the microscope • AML – Favorable Risk • • • – Intermediate Risk • – All abnormalities not in favorable or high risk categories, plus normal Poor Risk • • • • inv (16) t (8;21) t (15;17) Monosomy 5 or 7; 5q-; 7qt (9;22) Complex (3 or more abnormalities) ALL – Favorable risk • – Hyperdiploidy High Risk • • • t (1;19) t (9;22) t (4;11) Pre-TED Pre TED Form (1): Cyto data • First three AML cyto abnormalities are associated with favorable prognosis (AML/ETO, e.g., refers to the two genes involved in the leukemia) • AML with 11q23: often associated with previous topoisomerase inhib.-based inhib. based chemotherapy (MLL gene is located at 11q23); usually t(9;11) (p22;q23) Pre-TED Pre TED Form (2) • IIn the th pre-TED TED example l above, b Ph Ph+ refers f tto the cytogenetics; bcr refers to the detection of bcr/abl gene product product, usually by PCR or FISH Form 2100 Chimerism Studies • N Note t th thatt for f collection ll ti off chimerism data, PCR is not an option and should not be recorded under “other” Disease Status: FISH • For data purposes, FISH is a subset of cytogenetics (cellular level) • Molecular evidence would be PCR and similar The Future of Prognosticating Outcomes in Acute Leukemia • May be based on the molecular biology of the leukemia as ascertained by – PCR – FISH – Microarray data/gene profiling • More and more critical to understand the molecular basis as more targeted therapies b become available il bl – Anti-bcr/abl drugs: imatinib and 2nd generation drugs – Anti flt3 etc etc. Summary • A varietyy of chromosomal abnormalities can be characterized and described using cytogenetics • Non-random chromosomal alterations occur, can define the disease (e.g. APML), and can have important prognostic value • Not all genetic abnormalities can be seen using cytogenetic techniques (e (e.g. g normal cytogenetics in AML) • Newer techniques (polymerase chain reaction [PCR] flfluorescentt in-situ [PCR], i it h hybridization b idi ti [FISH]) can assist in searching for occult genetic aberrations Web References • http://www.slh.wisc.edu/wps/wcm/connect/e xtranet/cytogenetics/ t t/ t ti / • http://ghr.nlm.nih.gov/handbook/illustrations