Supplementary Information
advertisement

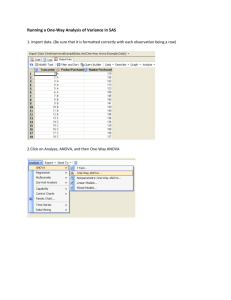
Supplementary information Statistics Statistical evaluation of the quantitative RT-PCR data for the three Aph-1 family members was performed by means of a univariate analysis of variance (ANOVA) with as dependent variable the normalized transcript levels and as fixed factors the genotypes (I/I, II/II and III/III) and the developmental time points (either ED or PND). For every time point, data were further analyzed for significant differences between the three genotypes using a one-way ANOVA and where appropriate a post hoc Bonferroni test. Per genotype, the statistical analysis for differences in transcript levels during development was performed by means of an independent samples T-test to compare the two embryonic time points, or using a one-way ANOVA and a post hoc Bonferroni test for the postnatal time points. A probability of P < 0.05 was considered statistically significant. For the longitudinal Western blot analyses, the data sets were statistically analyzed by means of a univariate ANOVA with as dependent variable the APPCTF/APP-FL levels and as fixed factors the genotypes and the developmental time points. Subsequent analysis using an independent samples T-test at every time point revealed significant differences between I/I and III/III rat tissues. Per rat line, a one-way ANOVA and where appropriate a post hoc Bonferroni analysis was used to identify differences between developmental time points. All statistical analyses were performed with the SPSS 12.0.1 software program (SPSS Inc., Chicago, Illinois, USA). Supplemental table 1 Statistical support for Figure 1 by means of a univariate ANOVA on longitudinal mRNA expression levels of Aph-1 family members in the hippocampus of I/I, II/II and III/III rats (significant if P < 0.05; NS: no significant differences). ED PND Aph-1b Aph-1aS Aph-1aL geno: F(2,19) = 57.6 geno: NS geno: NS ED: F(1,19) = 9.8 ED: F(1,21) = 12.6 ED: F(1,22) = 18.9 geno*ED: F(2,19) = 4.5 geno*ED: NS geno*ED: NS geno: F(2,81) = 242.3 geno: NS geno: NS PND: F(1,81) = 26.8 PND: F(1,86) = 64.1 PND: F(1,86) = 20.0 geno*PND F(1,81) = 3.2 geno*PND: NS geno*PND: NS Supplemental table 2 Since the univariate ANOVA for Figure 1 yielded statistical significance, we performed for the I/I, II/II and III/III rats over the embryonic time points an independent samples T-test and over the postnatal time points a one-way ANOVA and where appropriate a post hoc Bonferroni analysis. Significant differences between time frames are indicated (significant if P < 0.05; NS: no significant differences). ED PND independent one-way ANOVA Bonferroni samples T-test PND(0-4) < PND(22-100) I/I NS PND: F(7,27) = 17.7 PND(9-12) < PND(35-60) II/II NS PND: F(7,25) = 5.5 PND(0) < PND(22) Aph-1b PND(0-4) < PND(22-100) III/III ED13 < ED17 PND: F(7,26) = 17.2 PND(0-4) < PND(9) PND(12) < PND(22-35) Aph-1aS Aph-1aL I/I NS PND: F(7,27) = 19.1 PND0-60) < PND(100) II/II NS PND: F(7,27) = 17.9 PND(0-60) < PND(100) III/III NS PND: F(7,29) = 29.8 PND(0-60) < PND(100) I/I NS PND: F(7,27) = 11.6 PND(0-9) < PND(35-60) II/II NS PND: F(7,27) = 3.2 NS III/III NS PND: F(7,29) = 11.4 PND(0-12) < PND(35-100) Supplemental Table 3 Since the univariate ANOVA for Figure 1 yielded statistical significance, we performed for every developmental time point a comparison of the transcript levels of the I/I, II/II and III/III rats by means of a one-way ANOVA and where appropriate a post hoc Bonferroni analysis. Significant differences between genotypes are indicated (significant if P < 0.05; NS: no significant differences). 13 ED 17 0 4 9 12 PND 22 35 60 100 one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni one-way ANOVA Bonferroni Aph-1b Aph-1aS Aph-1aL geno: F(2,8) = 34.1 (I/I or II/II) < III/III geno: F(2,9) = 34.0 (I/I or II/II) < III/III geno: F(2,8) = 129.5 (I/I or II/II) < III/III geno: F(2,8) = 52.0 (I/I or II/II) < III/III geno: F(2,8) = 148.2 (I/I or II/II) < III/III geno: F(2,8) = 90.9 (I/I or II/II) < III/III geno: F(2,10) = 10.7 (I/I or II/II) < III/III geno: F(2,8) = 135.5 I/I < II/II < III/III geno: F(2,15) = 51.9 I/I < II/II < III/III geno: F(2,8) = 295.3 I/I < II/II < III/III NS NS NS NS NS NS NS NS NS NS - NS NS NS NS NS NS NS NS NS geno: F(2,8) = 5.2 NS Supplemental table 4 Statistical support for Figure 2 by means of a univariate ANOVA on longitudinal APP-CTF levels in the spinal cord and lung of I/I and III/III rats. Since the univariate ANOVA yielded statistical significance, we performed an independent samples T-test over the APP-CTF levels for every developmental time point, and for both I/I and III/III rats a one-way ANOVA and post hoc a Bonferroni analysis. Significant differences between genotypes or time frames are indicated (significant if P < 0.05; NS: no significant differences). spinal cord univariate ANOVA geno: NS PND: F(4,30) = 15.6 geno*PND: NS geno I/I III/III PND 2 9 13 35 60 independent samples T-test NS NS I/I > III/III I/I > III/III NS one-way ANOVA F(4,14) = 4.4 F(4,14) = 46.2 Bonferroni PND(2) < PND(35) PND(2-13) < PND35-60) lung univariate ANOVA geno: F(1,30) = 13.1 PND: F(4,30) = 22.7 geno*PND: F(4,30) = 3.4 geno I/I III/III PND 2 9 13 35 60 independent samples T-test NS NS NS I/I > III/III I/I > III/III one-way ANOVA F(4,14) = 21.3 F(4,14) = 4.4 Bonferroni PND(2-9) < PND(13-60) NS