Supplementary Information Histone modifications are associated
advertisement

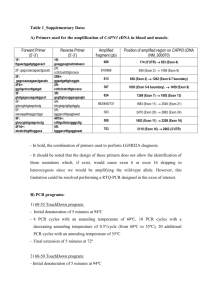
Supplementary Information Histone modifications are associated with transcript isoform diversity in normal and cancer cells Ondrej Podlaha1, Subhajyoti De2,3,4, Mithat Gonen5, and Franziska Michor1* 1Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute, and Department of Biostatistics, Harvard School of Public Health, Boston, MA 02215, USA. 2Department of Medicine, University of Colorado School of Medicine, Aurora, CO 80045, USA. 3Department of Biostatistics and Informatics, Colorado School of Public Health, Aurora, CO 80045, USA. 4Molecular Oncology Program, University of Colorado Cancer Center, Aurora, CO 80045, USA. 5Department of Epidemiology and Biostatistics, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA. *Author for correspondence. Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute, 450 Brookline Avenue, Boston, MA 02215, USA. Tel: 617 643 5045. Fax: 617 632 2444. Email: michor@jimmy.harvard.edu. *Author for correspondence. 0.19414 0.20887 0.21417 0.20439 0.22092 0.2197 0.20944 0.1689 0.18625 0.19439 0.19874 0.19932 0.19623 0.18684 0.18261 0.19586 0.19913 0.196 0.18104 0.1809 0.18183 0.06013 0.02652 -0.0103 -0.0042 0.03215 0.0492 0.06678 0.18245 0.19155 0.20042 0.1997 0.18191 0.18299 0.17865 -0.0931 -0.0849 -0.0807 -0.0637 -0.067 -0.0749 -0.077 0.18285 0.17339 0.1458 0.11141 0.17047 0.18304 0.18827 0.10298 0.13896 0.15884 0.15922 0.15337 0.14385 0.1176 Ct az H2 H3 0.13219 0.06034 0.01543 0.00439 0.05415 0.09123 0.13604 cf e2 e3 k7 9m e3 k3 6m H3 k2 7m H3 0.0018 -0.0333 -0.0551 -0.0508 -0.053 -0.0506 -0.0458 k2 7a H3 -0.0936 -0.0884 -0.0834 -0.07 -0.0709 -0.0798 -0.0825 c e1 k2 0m e3 k9 m e1 k9 m H3 k9 ac e3 H3 k4 m e2 H3 k4 m H3 k4 m H3 0.17302 0.1763 0.15372 0.12824 0.1946 0.19834 0.18888 H4 Average of Fisher Transformed Spearman rank correlations for normal cells Distance from exon -5kb -2kb -1kb 0kb +1kb +2kb +5kb H3 lincRNA TSSIR e1 Table S1. Association between the transcription start site inclusion rate (TSSIR) of lincRNAs and histone modification enrichment in normal cell lines. We analyzed six normal human cell lines (Gm12878, Hsmm, Huvec, H1hesc, Nhek, Nhlf) for the associations between the transcription start site inclusion rate (TSSIR) and histone modification enrichment for lincRNAs. Values represent the average of Fisher transformed Spearman rank correlations to enable direct comparison. Coefficients are color-coded, with red representing increasingly negative and green representing increasingly positive correlation. Distance from exon categories signifies a region relative to a given exon where histone enrichment was measured. 0kb represents a region within given exon boundaries, and 1kb, 2kb, and 5kb signify regions from the exon boundary either upstream (negative) or downstream (positive). 0.10297 0.11261 0.11568 0.08318 0.08444 0.07837 0.06618 Ct cf H2 az e2 e3 H3 k7 9m H3 k3 6m e3 H3 k2 7m H3 k2 7a c e1 H4 k2 0m e3 H3 k9 m e1 H3 k9 m H3 k9 ac e3 H3 k4 m e2 k4 m k4 m H3 Average of Fisher Transformed Spearman rank correlations for cancer cells Distance from exon -5kb -2kb -1kb 0kb +1kb +2kb +5kb H3 Protein coding Splicing e1 Table S2. Association between splicing exon inclusion rate (SEIR) of protein coding genes and histone modifications in cancer cell lines. We analyzed three cancer cell lines (Hepg2, Helas3, K562) for associations between splicing exon inclusion rate (SEIR) and histone modification enrichment for protein coding genes. Values represent the average of Fisher transformed Spearman rank correlations to enable direct comparison. Coefficients are color-coded, with red representing increasingly negative and green representing increasingly positive correlation. Distance from exon categories signifies a region relative to a given exon where histone enrichment was measured. 0kb represents region within given exon boundaries, and 1kb, 2kb, and 5kb signify regions from the exon boundary either upstream (negative) or downstream (positive). -0.1264 -0.1204 -0.0688 -0.1399 -0.159 -0.1433 -0.0498 -0.2367 -0.2231 -0.1773 -0.2532 -0.2615 -0.2579 -0.1907 -0.2353 -0.223 -0.1839 -0.2437 -0.2501 -0.2515 -0.1958 -0.1345 -0.1065 -0.0591 -0.1475 -0.1351 -0.112 -0.174 0.15088 0.16649 0.17715 0.13425 0.13922 0.14339 0.06073 -0.0553 -0.0666 -0.0887 -0.0555 -0.0551 -0.0692 -0.0396 0.07855 0.09038 0.09863 0.01208 0.00976 0.01514 0.00168 -0.1433 -0.1157 -0.0679 -0.1531 -0.1376 -0.1094 -0.1384 -0.2172 -0.2291 -0.243 -0.2317 -0.2432 -0.2588 -0.1557 0.33112 0.32191 0.3049 0.38082 0.36847 0.33757 0.28785 0.02958 0.05967 0.10801 -0.0347 -0.0212 0.00058 -0.0229 -0.2238 -0.2192 -0.1921 -0.225 -0.2353 -0.2356 -0.2164 -0.1341 -0.117 -0.0894 -0.1223 -0.1076 -0.0802 -0.1099 lincRNA TSSIR Average of Fisher Transformed Spearman rank correlations for cancer cells 0.18236 0.18136 0.15496 0.12096 0.19572 0.20292 0.20012 0.18413 0.19889 0.20463 0.20517 0.2186 0.21546 0.21656 0.18131 0.19575 0.20116 0.20588 0.20999 0.20723 0.20768 0.19036 0.20194 0.20348 0.20127 0.20923 0.20741 0.20573 0.03933 0.03041 -0.001 -0.0046 0.00853 0.0248 0.03642 -0.0621 0.0378 0.20156 -0.0472 0.01576 0.21458 -0.0427 -0.0178 0.21322 -0.0218 -0.0172 0.20345 -0.0474 -0.0354 0.21293 -0.0656 -0.0328 0.20882 -0.0629 -0.0132 0.20996 -0.0733 -0.0479 -0.0429 -0.0293 -0.0502 -0.0676 -0.0795 0.15204 0.0921 0.02366 0.01656 0.07038 0.11181 0.15723 0.17973 0.16921 0.15336 0.12869 0.17467 0.18386 0.19777 0.10571 0.14187 0.15004 0.14774 0.15045 0.13667 0.11818 cf Ct H2 az e2 e3 k7 9m H3 k3 6m e3 H3 k2 7m H3 H3 k2 7a c e1 k2 0m e3 H4 k9 m e1 H3 k9 m H3 k9 ac e3 H3 k4 m e2 H3 k4 m H3 H3 Distance from exon -5kb -2kb -1kb 0kb +1kb +2kb +5kb k4 m e1 Table S3. Association between transcription start site inclusion rate (TSSIR) of lincRNAs and histone modification enrichment in cancer cell lines. We analyzed three cancer cell lines (Hepg2, Helas3, K562) for associations between transcription start site inclusion rate (TSSIR) and histone modification enrichment for lincRNAs. Values represent the average of Fisher transformed Spearman rank correlations to enable direct comparison. Coefficients are color-coded, with red representing increasingly negative and green representing increasingly positive correlation. Distance from exon categories signify region relative to a given exon where histone enrichment was measured. 0kb represents region within given exon boundaries, and 1kb, 2kb, and 5kb signify regions from the exon boundary either upstream (negative) or downstream (positive). 0.1007 0.11348 0.1186 0.08401 0.08887 0.07812 0.07658 lincRNA Splicing Average of Fisher Transformed Spearman rank correlations for cancer cells 0.07083 0.05181 0.04643 0.04437 0.04117 0.05316 0.05464 0.04938 0.02422 0.01278 0.01107 0.01633 0.02978 0.02587 0.04611 0.02366 0.00786 0.00734 0.0091 0.01871 0.02135 0.0469 -0.0376 0.02734 -0.0216 0.01699 -0.0043 0.00202 -0.0154 0.01646 -0.0073 0.03418 0.00309 0.03263 -0.0077 -0.0029 0.00367 0.00968 0.00065 -0.0015 0.00943 -8E-08 -0.0234 -0.0289 -0.0146 -0.0057 -0.0134 -0.0066 -0.0058 0.06169 0.03241 0.0202 0.03141 0.02087 0.03846 0.04028 -0.0438 -0.0438 -0.0382 -0.0344 -0.0485 -0.0411 -0.0425 0.06375 0.04163 0.05165 0.06258 0.05638 0.07357 0.07824 Ct cf e2 az H2 H3 k7 9m e3 H3 k3 6m e3 k2 7m H3 H3 k2 7a c e1 k2 0m H4 e3 H3 k9 m e1 k9 m H3 k9 ac H3 e3 H3 k4 m e2 k4 m H3 H3 Distance from exon -5kb -2kb -1kb 0kb +1kb +2kb +5kb k4 m e1 Table S4. Association between splicing exon inclusion rate (SEIR) of lincRNAs and histone modifications in cancer cell lines. We analyzed three cancer cell lines (Hepg2, Helas3, K562) for associations between splicing exon inclusion rate (SEIR) and histone modification enrichment for protein coding genes. Values represent the average of Fisher transformed Spearman rank correlations to enable direct comparison. Coefficients are color-coded, with red representing increasingly negative and green representing increasingly positive correlation. Distance from exon categories signifies a region relative to a given exon where a histone enrichment was measured. 0kb represents region within given exon boundaries, and 1kb, 2kb, and 5kb signify regions from the exon boundary either upstream (negative) or downstream (positive). 0.05791 0.02593 0.01036 0.04083 0.00101 -0.0055 0.03466 -0.0096 -0.0103 0.0134 -0.0391 -0.0115 0.02072 -0.0205 -0.0056 0.04411 -0.0006 0.00556 0.0518 -0.0043 0.01127 Table S5. Association between histone modification enrichment and transcription start site inclusion rate. Correlations between transcription start site inclusion rate or splicing and enrichment of selected histone modifications in normal or cancer cell lines. The following correlations and cell line categories are: (A) transcription start site switching and normal cell lines, (B) splicing and normal cell lines, (C) transcription start site switching and cancer cell lines, and (D) splicing and cancer cell lines. Black dots represent median Spearman rank correlations between exon inclusion rate and given histone marks. All correlation coefficients were transformed using a Fisher’s transformation before plotting. Notches were calculated as æ IQR ö ±1.58´ ç ÷ where IQR stands for inter quartile range and n for sample size. Distances from è n ø exon represent genomic blocks of a given size from exon start (upstream regions) or exon end (downstream regions). A B C D Table S6. Top 20 ontology categories enriched among 840 candidate genes that showed a significant association between splicing exon inclusion rates and histone modification enrichment. GO term GO:0048583 GO:0051716 GO:0044763 GO:0009966 GO:0050794 GO:0065007 GO:0007165 GO:0044767 GO:0023051 GO:0032502 GO:0010646 GO:0050789 GO:0048522 GO:1902531 GO:0051128 GO:0019222 GO:0044699 GO:0006928 GO:0048518 GO:0009987 Description regulation of response to stimulus cellular response to stimulus single-organism cellular process regulation of signal transduction regulation of cellular process biological regulation signal transduction single-organism developmental process regulation of signaling developmental process regulation of cell communication regulation of biological process positive regulation of cellular process regulation of intracellular signal transduction regulation of cellular component organization regulation of metabolic process single-organism process cellular component movement positive regulation of biological process cellular process P-value 1.77E-10 2.53E-10 2.66E-10 5.79E-10 1.57E-09 3.93E-09 4.47E-09 4.93E-09 9.31E-09 1.13E-08 1.34E-08 1.34E-08 2.30E-08 4.11E-08 6.87E-08 7.50E-08 1.69E-07 2.00E-07 2.37E-07 2.57E-07 FDR q-value 2.11E-06 1.51E-06 1.05E-06 1.72E-06 3.74E-06 7.79E-06 7.60E-06 7.33E-06 1.23E-05 1.35E-05 1.45E-05 1.33E-05 2.10E-05 3.49E-05 5.45E-05 5.58E-05 1.19E-04 1.33E-04 1.49E-04 1.53E-04 Table S7. Leave-one-out cross validation summary statistics. Validation cell line Gm12878 H1hesc Hsmm Huvec Nhek Nhlf Helas3 Hepg2 K562 Mono Hmec Total Number Of Genes 29354 29354 29354 29354 29354 29354 29354 29354 29354 29354 29354 Number of Genes Assigned to Categories 2750 2745 2770 2696 2753 2786 2357 2720 2707 2519 2792 Number of Significant Genes 686 660 665 652 667 687 455 684 672 654 669 Number of Significant Cancer Genes 26 23 29 25 23 27 15 21 21 22 29 Module S1. Epigenetically aberrant regions in three cancer cell lines are enriched for oncogenes. Using the cancer gene consensus from COSMIC, we tested for oncogene enrichment in epigenetically aberrant regions of three cancer cell lines (Helas3, Hepg2, and K562) with regard to specific histone marks using the hypergeometric test. All p e2 H2 az 9m e3 H3 k7 6m e3 H3 k3 c 7a 7m H3 k2 e1 H3 k2 0m m e3 H4 k2 H3 k9 ac e3 H3 k9 m e2 H3 k4 m H3 k4 CellLine Helas3 0.06855 0.0916 0.0058 0.0231 0.0231 0.0231 0.0054 0.0684 0.8893 0.0003 3E-06 0.0054 Hepg2 0.00181 0.3192 0.61 0.0085 0.3566 NA 0.0003 0.3216 0.644 0.0018 0.0002 0.2707 K562 0.63255 0.0922 0.2228 0.0052 0.0052 0.9498 0.0922 0.0922 0.6325 0.0922 4E-05 0.6325 Ct Hypergeometric test p value H3 k4 cf m e1 values were corrected for multiple testing (FDR). Green highlights significantly enriched marks at the 5% FDR level.