Sample/Template - Springer Static Content Server
advertisement

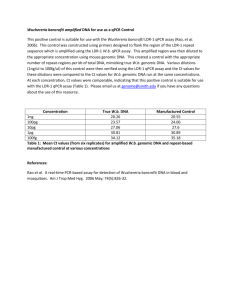
Table 4s MIQE list Sample/Template instruction details Source If cancer,was biopsy screened for adjacent normal tissue? LiquidN2/RNAlater/formalin If using samples>6months old Human genomic DNA and lambda DNA fresh/frozen/formalin Series of dilution of either human genomic DNA or lambda DNA were freshly prepared each time Human genomic DNA was extracted with EZ Spin Column Whole Blood Genome DNA Isolation Kit (Cat.# SK1261,Sangon). Lambda DNA was purchased from Sangon (Cat# SD0011). Not applied for this study Method of preservation Storage time (if appropriate) Handling Extraction method TriZol/columns RNA:DNA-free Concentration RNA:integrity Inhibition-free Intron-spanning primers/no RT control Nanodrop/ribogreen/microfluidics Microfluidics/3':5' assay Method of testing Assay optimisation/validation Accession number RefSeq XX_1234567 Amplicon details exon location, amplicon size Primer sequence even if previously published Probe sequence* In silico identify LNA or other substitutions BLAST/Primer-BLAST/m-fold -80 degree refrigerator The genomic DNA samples were stored at -80 degree within 6 months About 10ng/l Not applied for this study Not applied for this study Amplifications were not designed for any specific gene or DNA fragment. Primers were randomly designed. Human genome amplification: B9-10 (862 bp amplicon), A99-100 (803 bp amplicon), B17-18 (994 bp amplicon), A93-94 (967 bp amplicon); Lambda DNA amplification: Q7-8 (116 bp amplicon) Randomly designed primers using Primer Premier 5: B9 5’-AGGTAAAGGGGACTTTGTCTTGC-3’ B10 5’-CATTCTGCATGATGCGGTTATTA-3’ A99 5’-ACTGGGAAACTGTGACTGCTGC-3’ A100 5’-GACCTAACGTGGGACATAGAACAA-3’ B17 5’-ATTCCTTGCCATAGCCAACAGTA-3’ B18 5’-CGAGATTTCCTGCCCTAATCTTT-3’ A93 5’-GAACAGTCCAACAGCACTGAGTAAA-3’ A94 5’-AGCAGTGGGCCTTCTGTAAAAT-3’ Q7 5’-GGCTTGGCTCTGCTAACAC-3’ Q8 5’-TCATTCAACACCCGCACTAT-3’ Not applied for this study Not applied for this study empirical primer concentration/annealing temperature Priming conditions PCR efficiency Linear dynamic range Limits of detection oligo-dT/random/combination/target-specific dilution curve spanning unknown targets LOD detection/accurte quantification Intra-assay variation copy numbers not Cq RT/PCR Protocols Reagents Duplicate RT NTC NAC Positive control Data analysis Specialist software Statistical justification Transparent,validated normalisation fig7s detailed description, concentrations, volumes supplier, Lot number ΔCq Cq & melt curves ΔCq beginning:end of qPCR inter-run calibrators e.g., QBAsePlus e.g., biological replicates e.g., GeNorm summary All primers (2uM) were added 0.8ul for 12 ul PCR. Annealing temperatures were all taken as 56 degree. Target specific priming dilution curve At least to 10-6 dilution of the stock solution (10ng/ul) of the template DNA When the stock solution (10ng/ul) was diluted 106 fold and 0.5ul (~1х105 lambda DNA molecules, as in fig1 and fig2) was added into a 12-ul qPCR, the full repeatability can be realized both for SYBR Green I- and EvaGreen –based qPCR, but Evagreen performed better in general. The lowest concentrations for LOD detection were not further detected. Assay variation of EvaGreen –based qPCR is generally smaller than that of SYBR Green I-based qPCR in this study. One representative demonstration can be seen in the fig7s. Not applied for this study Thermal Cycler DiceTM Real Time System Software Biological replicates. Each set of PCR was repeated 2-4 times. Not applied for this study. This study didn’t quantify any gene. Only basic qPCR performance, such as melting-curves and amplification plots, was presented in order to demonstrate the enhancing effects of QDs for SYBR Green I- and EvaGreen –based qPCR. A B C D E Fig 7s Intra-assay or inter-assay variation of EvaGreen –based qPCR is generally smaller than that of SYBR Green I-based qPCR in this study. A) amplification plot (SYBR Green I, Cat#SY1020, Solarbio); B) melting curve (SYBR Green I); C) amplification plot (EVAGREEN, Cat#31000,Biotium); D) melting curve (EVAGREEN); E) Repeatability assay of EvaGreen –based qPCR with 10-2,10-3,10-5,10-6,10-7 dilutions of the template DNA. SYBR Green I-based qPCR had similar repeatability. PCR conditions: 94 oC-3min-(94 oC-20s-56 oC-32s-72 oC-24s)*45—72 oC-3min. TAKARA QPCR TP800 Primers Q3-4 (amplicon size 106bp) Q3 5’-CTAATAAGCCGATAGATAGCCAC-3’ Q4 5’-TACCTTTCCGCCATAACTGT-3’ 12ul qPCR system: 12 ul reaction 2 *PCR buffer (NPK02) 1 ul 6.0uL ddH20 2.2uL Dye (20х) 0.6uL QDs 1.5 ul Taq 0.2 ul Human genomic DNA 0.5uL Primer 3-4 (10ng/ul) -1 Template dilution 0.5 0.5*10 Plot line color red Bright blue 0.5*10 green -2 0.5*10 yellow -3 0.5*10 pink -4 0.5*10 blue -5 0.5*10-6 2.7*10-7 orange black