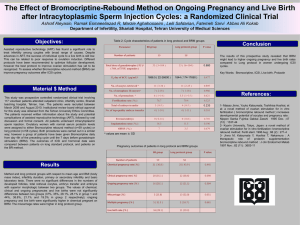
hep26207-sup-0001-supinfo
advertisement

Tian WF et al: Brahma related gene 1 (Brg1) bridges epigenetic regulation of proinflammatory cytokine production to steatohepatitis Supplemental Materials and Methods Supplementary figures: 5 Supplementary tables: 1 Materials and Methods Plasmids and RNA interference Promoter luciferase fusion constructs for IL-6, MCP-1, IL-8, iNOS, Brg1 and Brm expression constructs, and short hairpin RNA (shRNA) construct targeting Brg1 or Brm have been described previously (1-5). Small interfering RNA (siRNA) sequences were as follows: for NF-B/p65, UGACGUAAAGGGAUAGGGC; for Brg1, GCCCATGGAGTCCATGCAT; and for Brm, AAGTCCTGGACCTCCAAGTGT. Animals To induce steatohepatitis, 8 week-old male db/db mice (Jackson Laboratory) were fed a methionine- and choline-deficient (MCD) diet (A02082002B, Research Diets) for 4 consecutive weeks. In certain experiments, these mice were injected via tail vein purified lentiviral particles (1X109 MOI) that carry short hairpin RNA (shRNA) targeting both Brg1 and Brm (GCUGGAGAAGCAGCAGAAG) at week 1 and week 3. Alternatively, 8 week-old male C57/BL6 mice were fed a high fat high carbohydrate (HFHC) diet (D12492, Research Diets) for 16 consecutive weeks (6). Blood triglyceride (TG) and alanine aminotransferase (ALT) levels were measured as described before (7). A third NASH model was made in which 6-8 week-old male Apoe-/- mice (Jackson Laboratory) were fed with a high fat diet (D12079, Research Diets) for 12 weeks. Histology Paraffin sections were stained with hematoxylin and eosin (Sigma), oil red O (Sigma), or picrosirius red (Sigma) according to standard procedures. Parallel sections were stained for cell surface markers. Briefly, the sections were blocked with 10% normal goat serum for 1 hour at room temperature and then incubated with anti-CD3 (BD Bioscience), anti-CD45 (Abcam), or anti-F4/80 (Abcam) antibodies. For ROS measurement, frozen liver sections were stained with dihydroethidium (DHE, Sigma). Slides were visualized under and photographed by an Olympus IX-70 microscope. Histology data were analyzed using Image Pro (Media Cybernetics). Hepatic pathology was scored by two independent investigators as described before (8). B A VEC 5 PA OA 4.5 HepG2 2.5 HepaRG Brg1 Brg1 Brm Brm -actin -actin 4 Relative mRNA levels Relative mRNA levels 2 HepG2 HepaRG 1.5 1 3.5 VEC OA 3 PA VEC OA PA 2.5 2 1.5 0.5 1 0.5 0 Brg1 0 Brm Brg1 Brm C HepG2 HepaRG 1.5 1 Relative mRNA levels Relative mRNA levels 2 2.5 2 1.5 1 0.5 0.5 0 Glu (h) Mannitol (h) 0 3 0 Glu (h) Manitol (h) 6 12 24 24 0 6 12 24 24 3 Brg1 mRNA 1.8 3 2.5 Relative mRNA levels Brm mRNA 3.5 1.8 1.6 1.6 1.4 1.4 Relative mRNA levels Brg1 mRNA 3 1.2 1 0.8 0.6 1.2 1 0.8 0.6 0.4 0.4 0.2 0.2 0 Glu (h) 0 3 6 Brm mRNA 0 Glu (h) 0 12 3 6 12 D Brm -actin Glucose (h) 3 0h 3h 6h 12h 2.5 2 HepaRG Brg1 1.5 Brm 1 -actin 0.5 0 Glucose (h) Brg1 Brm E F HepG2 2.5 3 1.4 Primary murine hepatocyte 1.2 HepG2 2.5 1.5 1 1 Relative mRNA levels Relative mRNA levels Relative mRNA levels 2 0.8 0.6 0.4 0.5 Brg1 Brm 0 PA E2 1.5 1 0.5 0.2 0 PA E2 2 Brg1 Brm 0 Glucose E2 Brg1 Brm Relative protein levels (random units) Brg1 Relative protein levels (random units) 3 HepG2 0h 3h 6h 12h 2.5 2 1.5 1 0.5 0 Brg1 Brm G H 1.4 Apoe-/- AL (N=5) Apoe-/- HFD (N=5) 4 * AL Brm 1 -actin 0.8 0.6 Apoe-/- AL (N=4) Apoe-/- HFD (N=4) * 3.5 Brg1 1.2 Relative mRNA levels HFD Relative protein levels (random units) 1.6 3 2.5 2 * 1.5 1 0.5 0 Brg1 0.4 Brm 0.2 0 Brg1 Brm Figure S1: (A, B) HepG2 and HepaRG cells were treated with a vehicle (VE), palmitate (0.4mM), or oleate (0.4mM) for 6 hours. mRNA (A) and protein (B) levels of Brg1 and Brm were assessed by qPCR and Western. (C, D) HepG2 and HepaRG cells were treated with or without glucose (35mM). Cells were harvested at indicated time points to probe for mRNA (A) and protein (B) levels of Brg1 and Brm. (E, F) HepG2 cells or Primary murine hepatocytes were treated with PA (E) or glucose (F) in the presence or absence of 17-estradiol (E2, 10-8~10-6M). Expression of Brg1 and Brm was measured by qPCR. (G, H) 6-8 week-old male Apoe-/- mice were fed with a high fat diet (HFD) for 12 weeks. Liver homogenates were probed for mRNA (G) and protein (H) expression of Brg1 and Brm. A AL HFHC Brg1 IL-6 promoter Brm Brg1 MCP-1 enhancer Brm B IL-1 ChIP IL-6 ChIP 8 VEC PA OA 4.5 7 10 6 5 4 3 4 Relative enrichment Relative enrichment Relative enrichment MCP-1 ChIP 5 12 8 6 4 3.5 3 2.5 2 1.5 2 1 2 1 0.5 0 Brg1 0 Brm Brg1 0 Brm Brg1 Brm C 2 IL-1 intronic ChIP 3 IL-6 intronic ChIP 0h 3h 6h 9h 1.8 2.5 1.4 Relative enrichment Relative enrichment 1.6 1.2 1 0.8 0.6 0.4 2 1.5 1 0.5 0.2 0 0 Brg1 Brm Brg1 Brm D 5 5 IL-1 ChIP 9 IL-6 ChIP MCP-1 ChIP 8 4 3 2 3 2 1 1 0 0 VEC PA PA+E2 low PA+E2 high 7 Relative enrichment Relative enrichment Relative enrichment 4 6 5 4 3 2 1 Brg1 Brm 0 Brg1 Brm Brg1 Brm Figure S2: (A) Nuclear proteins were extracted from the livers of NASH mice. DNA affinity pull-down assay was performed as described under Materials and Methods. (B) HepG2 cells were treated with a vehicle (VE), palmitate (0.4mM), or oleate (0.4mM) for 6 hours. ChIP assays were performed with indicated antibodies. (C) HepG2 cells were treated with or without palmitate. ChIP assays were performed with indicated antibodies. Precipitated DNA was amplified by primers spanning the intronic region of the indicated gene. (D) HepG2 cells were treated with palmitate in the presence or absence of 17estradiol (E2 low, 10-7M; E2 high, 10-6M). ChIP assays were performed with indicated antibodies. A Relative mRNA levels 1 sip65 p65 mRNA SCR 1.2 p65 0.8 -actin 0.6 0.4 0.2 0 SCR sip65 B siBrm Relative mRNA levels 1 siBrg1 SCR siBrg1 siBrm 1.2 SCR 1.4 Brg1 0.8 Brm 0.6 -actin 0.4 0.2 0 Brg1 Brm Figure S3: (A) HepG2 cells were transfected with indicated siRNA. NF-B/p65 levels were measured by qPCR and Western. (B) HepG2 cells were transfected with indicated siRNA. Brg1 and Brm levels were measured by qPCR and Western. 1.4 1.2 * * * * Relative mRNA levels * SCR+V SCR+PA siBrg1/Brm+PA 1 0.8 0.6 0.4 0.2 0 Brg1 Brm IL-1 IL-6 TNF- Figure S4: Murine primary hepatic cells were infected with lentivirus carrying shRNA targeting Brg1/Brm or SCR followed by treatment with palmitate (PA) or vehicle (V). Expression levels of indicated genes were measured by qPCR. A AL+SCR MCD+SCR MCD+shBrg1/Brm B 0.7 Relative mRNA levels 0.6 db/db+SCR MCD db/db+shBrg1/Brm MCD * * 0.5 0.4 0.3 0.2 0.1 0 Col1a1 Col1a2 Figure S5: Lentiviral particles carrying shRNA targeting Brg1/Brm or SCR were injected via tail vein as described under Materials and Methods. (A) ROS levels were measured by DHE staining. (B) Hepatic collagen type I levels were measured by qPCR. N=3 Table I: ChIP Real-time qPCR primers Gene name Human IL-1 Human IL-6 Human MCP-1 Mouse Il-1 Mouse Il-6 Mouse Mcp-1 Primer sequences Forward: 5’-CTGTGTGTCTTCCACTTTGTCCC-3’ Reverse: 5’-TGCATTGTTTTCCTGACAATCG-3’ Forward: 5’-CTTCGTGCATGACTTCAGCTTT-3’ Reverse: 5’-CGTCCTTTAGCATGGCAAGAC-3’ Forward: 5’-CCCATTTGCTCATTTGGTCTCAGC-3’ Reverse: 5’-GCTGCTGTCTCTGCCTCTTATTGA-3’ Forward: 5’-TTCGATGCTGCCCTTTATCT-3’ Reverse: 5’-AGGCAGGTGGGTACTCATGTA-3’ Forward: 5’-GACATGCTCAAGTGCTGAGTCAC-3’ Reverse: 5’-AGATTGCACAATGTGACGTCG-3’ Forward: 5’-CGTGGGAAAATCCAGTATTTTAATG-3’ Reverse: 5’-CAAATGTATCACCATGCAAATATGC-3’ References: 1. Hoffmann E, Ashouri J, Wolter S, Doerrie A, Dittrich-Breiholz O, Schneider H, Wagner EF, et al. Transcriptional regulation of EGR-1 by the interleukin-1JNK-MKK7-c-Jun pathway. J Biol Chem 2008;283:12120-12128. 2. Ueda A, Ishigatsubo Y, Okubo T, Yoshimura T. Transcriptional regulation of the human monocyte chemoattractant protein-1 gene. Cooperation of two NFkappaB sites and NF-kappaB/Rel subunit specificity. J Biol Chem 1997;272:3109231099. 3. Zhou J, Zhang M, Fang H, El-Mounayri O, Rodenberg JM, Imbalzano AN, Herring BP. The SWI/SNF chromatin remodeling complex regulates myocardininduced smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol 2009;29:921-928. 4. Crosby MB, Svenson J, Gilkeson GS, Nowling TK. A novel PPAR response element in the murine iNOS promoter. Mol Immunol 2005;42:1303-1310. 5. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550-553. 6. Kohli R, Kirby M, Xanthakos SA, Softic S, Feldstein AE, Saxena V, Tang PH, et al. High-fructose, medium chain trans fat diet induces liver fibrosis and elevates plasma coenzyme Q9 in a novel murine model of obesity and nonalcoholic steatohepatitis. Hepatology 2010;52:934-944. 7. Witek RP, Stone WC, Karaca FG, Syn WK, Pereira TA, Agboola KM, Omenetti A, et al. Pan-caspase inhibitor VX-166 reduces fibrosis in an animal model of nonalcoholic steatohepatitis. Hepatology 2009;50:1421-1430. 8. Yu J, Ip E, Dela Pena A, Hou JY, Sesha J, Pera N, Hall P, et al. COX-2 induction in mice with experimental nutritional steatohepatitis: Role as proinflammatory mediator. Hepatology 2006;43:826-836.