Supplementary Table 1
advertisement
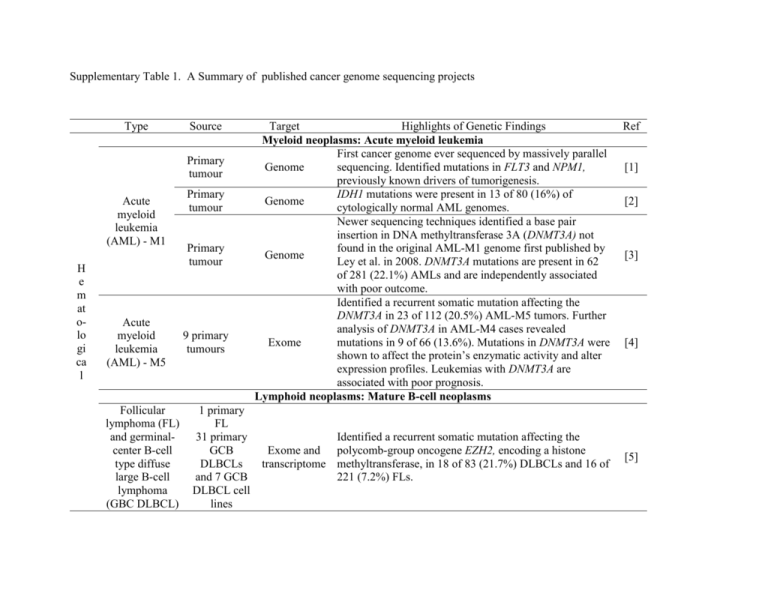
Supplementary Table 1. A Summary of published cancer genome sequencing projects Type Source Primary tumour Acute myeloid leukemia (AML) - M1 H e m at olo gi ca l Acute myeloid leukemia (AML) - M5 Follicular lymphoma (FL) and germinalcenter B-cell type diffuse large B-cell lymphoma (GBC DLBCL) Primary tumour Primary tumour 9 primary tumours Target Highlights of Genetic Findings Myeloid neoplasms: Acute myeloid leukemia First cancer genome ever sequenced by massively parallel Genome sequencing. Identified mutations in FLT3 and NPM1, previously known drivers of tumorigenesis. IDH1 mutations were present in 13 of 80 (16%) of Genome cytologically normal AML genomes. Newer sequencing techniques identified a base pair insertion in DNA methyltransferase 3A (DNMT3A) not found in the original AML-M1 genome first published by Genome Ley et al. in 2008. DNMT3A mutations are present in 62 of 281 (22.1%) AMLs and are independently associated with poor outcome. Identified a recurrent somatic mutation affecting the DNMT3A in 23 of 112 (20.5%) AML-M5 tumors. Further analysis of DNMT3A in AML-M4 cases revealed Exome mutations in 9 of 66 (13.6%). Mutations in DNMT3A were shown to affect the protein’s enzymatic activity and alter expression profiles. Leukemias with DNMT3A are associated with poor prognosis. Lymphoid neoplasms: Mature B-cell neoplasms 1 primary FL 31 primary Identified a recurrent somatic mutation affecting the GCB Exome and polycomb-group oncogene EZH2, encoding a histone DLBCLs transcriptome methyltransferase, in 18 of 83 (21.7%) DLBCLs and 16 of and 7 GCB 221 (7.2%) FLs. DLBCL cell lines Ref [1] [2] [3] [4] [5] Activated B-cell like diffuse large B-cell lymphoma (ABC DLBCL) 4 primary tumours Multiple myeloma (MM) 38 primary tumours Chronic lymphocytic leukemia (CLL) 4 primary tumours Hairy-cell leukemia (HCL) Primary tumor Hodgkin lymphoma 2 cancer cell lines Identified a single mutation, L265P, in MYD88 that was present in 111 of 382 lymphomas (29%). Additionally Transcriptom L265P was present in 9% of mucosa-associated lymphod e tissue lymphomas. Analysis of this mutation demonstrated a gain-of-function promoting cell survival through the NF-kB and JAK-STAT3 pathway. Analysis of 23 multiple myeloma genomes as well as 16 multiple myeloma exomes (including one that was analysed by both methods) revealed mutations involving Genome and genes that regulate: RNA processing, protein homeostasis, exome the NF-kB pathway, gene imprinting and the coagulation cascade. Additionally, mutations in BRAF and in noncoding regions were discovered. Identified 45 genes with mutations in protein-coding sequences. Analysis of these 45 genes in an additional 363 CLL patients identified 4 genes that are recurrently mutated: NOTCH1, MYD88, XPO1 and KLHL6. Genome Mutations identified in NOTCH1 and MYD88 are activating mutations present in 31/255 (12.2%) and 9/310 (2.9%) respectively. Mutations in NOTCH1 are associated with decreased overall survival. Exome sequencing identified 5 nonsynonymous mutations one of which was BRAF V600E. An additional 47 patients with HCL were examined. All had BRAF V600E Exome mutations and virtually all were present in the original clone. None were found in 195 patients with other peripheral B-cell lymphomas and leukemias. Lymphoid neoplasms: Hodgkin lymphoma Identified a gene fusion involving CIITA that was found to be recurrent in 29 of 77 B-cell lymphomas (38%) and 8 of Transcriptom 55 (15%) classical Hodgkin lymphomas. CIITA gene e fusions result in down-regulation of surface HLA class II expression and up-regulation of ligands of PDL1/2 [6] [7] [8] [9] [10] Br ea st an d O va ria n Lobular breast cancer Lung metastasis Basal-like breast cancer Primary tumour, brain metastasis and mouse xenograft [11] Genome Point mutations were predominantly C>T/G>A transitions. Comparing the mutational spectrum of the primary tumour with that of the metastases and xenograft, suggested that all the mutations necessary for metastases were already present in the primary tumour. [12] Triple negative breast cancer 2 primary tumours and liver metastasis Copy number variation of a singlenucleus for 100 cells of each tumour Phylogenetic reconstruction demonstrated that tumour progression is characterized by a “punctuated clonal evolution”. Findings also show that metastatic potential is achieved in the late stages of tumour evolution. [13] Granuolsa-cell tumour (GCT) 4 primary tumours Transcriptom e 8 primary tumours Exome Ovarian clearcell carcinoma (OCCC) L un g allowing the tumour cell to escape immune detection. Five mutations (in ABCB11, HAUS3, SLC24A4, SNX4 and Genome and PALB2) were prevalent in both the primary and the transcriptome metastatic tumour. RNA-editing events contribute to the transcriptional variation of lobular breast cancer. Small-cell lung cancer 18 OCCCs and 1 OCCC cell line NCI-H209 cell line Identified a single recurrent mutation 402C>G (C134W) in FOXL2 in all 4 GCTs. Sequencing of an additional 89 GCTs revealed the same mutation in 86 (97%). Identified PPP2R1A and ARID1A mutations in two tumours. Resequencing 42 OCCCs revealed PPP2R1A mutations were present in 7% and ARID1A mutations in 57% of OCCCs. ARID1A mutations were present in 55 of 119 (46%) Exome and ovarian clear-cell carcinomas, and 10 of 33 (30%) transcriptome endometrioid carcinomas. Genome Tabocco carcinogens predominantly cause G>T/C>A transversions, preferentially at methylated CpGs. Mutational signatures showed evidence of transcriptioncoupled nucleotide excision repair as well as expression- [14] [15] [16] [17] M el an o m a Br ai n Non-small-cell lung cancer Primary tumour Malignant pleural mesothelioma Primary tumour Melanoma COLO-829 cell line, a malignant cell line Uveal Melanoma class 2 2 tumours with monosomy 3 Glioblastoma multiforme 21 primary tumours Medulloblastom a (MB) 17 primary tumours, 4 xenografts, 1 cell line coupled repair. The mutational signature was similar to that observed in small-cell lung cancer. The predominant point mutations Genome were G>T/C>A transversions and in methylated CpGs with evidence of transcription-coupled repair. A DPP10 deletion was identified in the primary tumour. Resequencing of this gene in 53 tumour samples was Genome detected in 31 (55%) and showed loss of DPP10 is associated with a poorer survival. The mutational imprint left by UV damage is characterized predominantly by C>T/G>A transitions, particularly between two adjacent pyrimidines. These Genome mutations occur more frequently at the 3’ base of a pyrimidine dinucleotide and in CpG dinucleotides. There was also evidence for transcription-coupled nucleotide excision repair. Both tumours contained inactivating mutations in BRCA1-associated protein 1 (BAP1) at 3p21.1. An additional 26 of 31 (84%) class 2 (high metastatic risk) UMs showed mutations in BAP1, while only 1 of 26 class Exome 1 (low metastatic risk) UMs contained BAP1 mutations. Additionally, one patient had a germline BAP1 mutation, suggesting that germline mutations in BAP1 could be a novel cancer predisposition gene. BAP1 regulates a number of genes involved in metastatic transformation. Mutations in IDH1 were found in 18 of 149 (12%) GBMs Exome and and all affect amino acid R132. Mutations affecting R132 transcriptome are associated with a better prognosis. The number of nonsilent (nonsynonymous, missense, nonsense, indels or splice site) mutations per Exome medulloblastoma, a childhood cancer, was only 8.3, five to ten times fewer mutations compared to adult solid tumours. The discovery of recurring mutations in MLL2 [18] [19] [20] [21] [22] [23] 7 metastic MBs R en al G as tr oint es tin al Clear cell renal carcinoma (ccRCC) 7 primary tumours Colorectal cancer 3 cell lines and 8 xenografts all derived from liver metastases Pancreatic adenocarcinoma Exome Exome 24 primary tumours Exome 24 primary tumours Exome Primary tumour and metastasis from thirteen patients and MLL3, genes regulating transcription and chromatin remodeling, suggests that MB is caused by mechanisms that subvert normal brain development, explaining its rarity in adult populations. Identified truncating mutations in PBRM1. Sequencing of an additional 257 renal cell carcinomas identified truncating mutations in 88 (34%). PBRM1 maps to 3p, the same position as tumour suppressor genes, VHL and SETD2. This may be why 3p loss-of-heterozygosity is commonly seen in ccRCC. IDH1 mutations affecting R132 were overlooked as driver mutations, however they were retrospectively recognized after discovering similar mutations in glioblastoma multiforme (Parsons 2008) Grouped mutated genes into 69 gene sets of which 31 could be further grouped into 12 core signalling pathways. Expression data of these 31 gene sets suggest that they contribute to pancreatic tumourigenesis. Identified one pancreatic tumor, which harbored both a germline and somatically acquired deleterious mutation in PALB2. Sequencing PALB2 in 96 patients with familial pancreatic cancer identified 3 patients with germline PALB2 truncating mutations, while none were found in 1084 controls. Genomic rearrangemen “Fold-back inversions” are a distinct pattern of genomic ts by paired- instability in pancreatic cancer indicative of telomere end erosion and dysregulation of the G1-to-S transition. sequencing [24] [25] [26] [27] [28] Pro stat e Primary tumour and metastases from seven patients Genome Pancreatic neuroendocrine tumour (PanNET) 10 sporadic PanNETs Exome Hepatocellular carcinoma (HCC) 1 primary tumour Genome and exome Prostate 7 tumours Genome Analysis of the genome of pancreatic cancers at various spatial and temporal stages of its evolution demonstrate that it is initiated by a single parental clone, from which progressor mutations accumulate and drive clonal evolution. Estimated time from the initiated tumour cell to parental clone is 11.7 years, while another 3.4 years elapses before subclones with metastatic potential are achieved. This suggests that metastatic cells arise late in tumour development. Identified and validated mutations in 58 PanNETs in chromatin remodeling genes (MEN1 in 44% and either DAXX or ATRX in 43%) as well as mutations involving the mTOR pathway (14%). Whole genome sequencing of a hepatitis C virus induced HCC revealed a predominance of T>C/A>G transitions with evidence of transcription-coupled repair. Exome sequencing of the same tumour sample at a higher sequence depth revealed sub-clones with mutations in the tumour suppressor gene, TSC1. Complex genomic rearrangements in prostate cancer are characterized by balanced translocations. Furthermore, a single closed chain of rearrangements can disrupt multiple known cancer genes. [29] [30] [31] [32] Supplementary Table 1. References 1. Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK: DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008, 456(7218):66-72. 2. Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ: Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009, 361(11):1058-1066. 3. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O'Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK: DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010, 363(25):2424-2433. 4. Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ, Chen Z, Chen SJ: Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 2011, 43(4):309-315. 5. Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA: Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42(2):181-185. 6. Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, Shaffer AL, Romesser P, Wright G, Powell J, Rosenwald A, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Staudt LM: Oncogenically active MYD88 mutations in human lymphoma. Nature 2011, 470(7332):115-119. 7. Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR: Initial genome sequencing and analysis of multiple myeloma. Nature 2011, 471(7339):467-472. 8. Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, Bassaganyas L, Baumann T, Juan M, López-Guerra M, Colomer D, Tubío JM, López C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernández JM, Puente DA, Freije JM, Velasco G, Gutiérrez-Fernández A, Costa D, Carrió A, Guijarro S, Enjuanes A, Hernández L, Yagüe J, Nicolás P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjosé S, Piris MA, de Alava E, Miguel JS, Royo R, Gelpí JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigó R, Bayés M, Heath S, Gut M, Klatt P, Marshall J, Raine K, Stebbings LA, Futreal PA, Stratton MR, Campbell PJ, Gut I, López-Guillermo A, Estivill X, Montserrat E, López-Otín C, Campo E: Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; advance online publication. 9. Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, Pucciarini A, Bigerna B, Pacini R, Wells VA, Sportoletti P, Pettirossi V, Mannucci R, Elliott O, Liso A, Ambrosetti A, Pulsoni A, Forconi F, Trentin L, Semenzato G, Inghirami G, Capponi M, Di Raimondo F, Patti C, Arcaini L, Musto P, Pileri S, Haferlach C, Schnittger S, Pizzolo G, Foà R, Farinelli L, Haferlach T, Pasqualucci L, Rabadan R, Falini B: BRAF mutations in Hairy-cell leukemia. N Engl J Med 2011,364(24):2305-2315. 10. Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, Johnson NA, Zhao Y, Telenius A, Neriah SB, McPherson A, Meissner B, Okoye UC, Diepstra A, van den Berg A, Sun M, Leung G, Jones SJ, Connors JM, Huntsman DG, Savage KJ, Rimsza LM, Horsman DE, Staudt LM, Steidl U, Marra MA, Gascoyne RD.: MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011;advance online publication. 11. Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S: Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 2009, 461(7265):809-813. 12. Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O'Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER: Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010, 464(7291):999-1005. 13. Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M: Tumour evolution inferred by single-cell sequencing. Nature 2011, 472(7341):90-94. 14. Shah SP, Köbel M, Senz J, Morin RD, Clarke BA, Wiegand KC, Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E, Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y, Zeng T, Glover JN, Vanderhyden B, Zhao C, Parkinson CA, Jimenez-Linan M, Bowtell DD, Mes-Masson AM, Brenton JD, Aparicio SA, Boyd N, Hirst M, Gilks CB, Marra M, Huntsman DG: Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009, 360(26):2719-2729. 15. Jones S, Wang TL, Shih IeM, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N: Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010, 330(6001):228231. 16. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, HeraviMoussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG: ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med 2010, 363(16):1532-1543. 17. Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordoñez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA, Campbell PJ: A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010, 463(7278):184-190. 18. Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, Ha C, Johnson S, Kennemer MI, Mohan S, Nazarenko I, Watanabe C, Sparks AB, Shames DS, Gentleman R, de Sauvage FJ, Stern H, Pandita A, Ballinger DG, Drmanac R, Modrusan Z, Seshagiri S, Zhang Z: The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 2010, 465(7297):473477. 19. Bueno R, De Rienzo A, Dong L, Gordon GJ, Hercus CF, Richards WG, Jensen RV, Anwar A, Maulik G, Chirieac LR, Ho KF, Taillon BE, Turcotte CL, Hercus RG, Gullans SR, Sugarbaker DJ: Second Generation Sequencing of the Mesothelioma Tumor Genome. PLoS ONE 2010, 5(5):e10612. 20. Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR: A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 2010, 463(7278):191-196. 21. Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM: Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330(6009):1410-1413. 22. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW: An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321(5897):1807-1812. 23. Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE: The genetic landscape of the childhood cancer medulloblastoma. Science 2011, 331(6016):435-439. 24. Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA: Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011, 469(7331):539-542. 25. Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE: The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314(5797):268-274. 26. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW: Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321(5897):1801-1806. 27. Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, Iacobuzio-Donahue CA, Maitra A, Parmigiani G, Kern SE, Velculescu VE, Kinzler KW, Vogelstein B, Eshleman JR, Goggins M, Klein AP: Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 2009, 324(5924):217. 28. Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA: The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010, 467(7319):1109-1113. 29. Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA: Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467(7319):1114-1117. 30. Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N: DAXX/ATRX, MEN1, and mTOR Pathway Genes Are Frequently Altered in Pancreatic Neuroendocrine Tumors. Science 2011, 331(6021):1199 -1203. 31. Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T et al: High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet 2011, 43(5):464-469. 32. Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C et al: The genomic complexity of primary human prostate cancer. Nature 2011, 470(7333):214-220.


