doc
advertisement

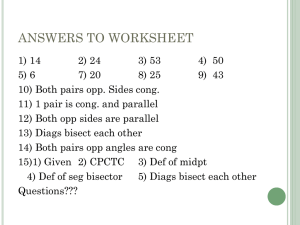
Lesson Notes Gene Regulation and Development: Phage —Handout 1 -Choose between lytic and lysogenic pathways -Lytic pathway -Immediate Early transcription—0-2’ after infection -Regulatory proteins -2 promoters—PL and PR -2 genes transcribed—N and cro -2 terminators—tL and tR1 -Delayed Early transcription (N-dependent)—2’-5’ -Replication of phage DNA -Genes transcribed going left from N = cIII, red, xis, int -Genes transcribed going right from cro = cII, O, P, Q -Most important genes = O,P -Needed for DNA synthesis -N protein allows reading through terminators of Immediate Early transcription -Either anti-terminator or activator of unknown promoter placed to the left of tL and to the right of tR1 -If activation, then IE RNA and DE RNA should be distinct; or if anti-termination, then DE RNA should be extension of IE RNA -If activation, then DE RNA might not depend on PL and PR; or if anti-termination, then DE RNA depend on initiation at PL and PR -Biochemically shown that N is anti-terminator -If supply N to PR-PL- cell and N is activator, then DE transcription should occur—NOT WORK -If supply N to lysogenic cell with cI present and N is activator, then DE transcription should occur—NOT WORK -Late transcription (Q-dependent)—5’-10’ -Make phage components and lyse cell -PR’ tR’ -Q allows PR’ to transcribe -Q is necessary for late transcription -Anti-terminator protein of tR’ -Since DNA is circular, A through J are transcribed -Code for head and tail proteins of phage particles -Aspects of lytic pathway -Transcriptional (regulatory) cascade -IE N Q late transcripts -N and Q are anti-terminators which act as positive regulators -N is a RNA binding protein -Interacts with RNAP as RNAP passes through nutL and nutR in mRNA 1 - nutL and nutR are secondary binding within mRNA -Stops RNAP from recognizing terminators -TAT protein of HIV like N -Q recognizes a DNA site which overlaps with PR’ -Interacts with RNAP to read through terminators -Lysogenic pathway -Clear plaques = lysogenization mutants -3 mutants found—cI, cII, and cIII -Cross streak each and if they complement each other, then turbid plaques form in intersection of cross cIcIIcIIIcI + + cII + cIII-+ = turbid plaques = complementation -- = clear plaques = no complementation -What mutant genes code for -cI = repressor -Activates PRM (promoter for repressor maintenance) while represses PL and PR -Leftward transcription of cI from PRM -Initially transcribes leftward from PRE (promoter for repressor establishment) -PRE is a weak promoter, therefore needs activator -cII = DNA binding protein -Activate PRE transcription of cI -Activate Pint which codes for integrase -Easily degraded -cIII = inhibitor of cII break down -Stabilize cII -cII and cIII are delayed early genes -Both need N -How does integration occur? -cII and cIII turn on PRE, which transcribes cI -Repressor (cI) binds to OR and OL, halting lytic pathway after Immediate Early transcription -Halts transcription of N; therefore, halts transcription of cII and cIII -PRM continues repressor transcription with positive activation by cI after PR and PL are repressed cI OR3 OR2 OR1 cro PRM PR -Repressor is able to bind to 3 sites, between PRM and PR—OR1, OR2, and OR3 in that order—Handout 2 Fig, 2.1, 2.2 2 -When bound to OR1, shut down PR by preventing RNAP to bind -Overlaps –10 region of PRE -Binds to OR2 at approximately the same time due to stabilization of binding to DNA through protein-protein interactions—Handout 2 Fig. 4.18 -When bound to OR2, it touches RNAP on PRM -OR2 is 1 bp closer to PR than to PRM—allows for different interactions between cI and RNAP -Acts as negative regulator of PR -Overlaps 3 bp of –35 region of PR by 1 bp, which prevents RNAP from binding -Acts as positive regulator of PRM—Handout 2 Fig. 1.16 -Interacts with -subunit of RNAP at –35 region of PRM -Overlaps -35 region by 2 bp -When bound to OR3, there is too much repressor present and halts transcription of cI until levels decrease enough so that not bound to OR3 anymore -Overlaps with –35 region of PRM -Allows for reversibility of repression -Needed for induction of prophage -Aspects of lysogenic pathway + -N cII, cIII PRE cI PRM -Regulatory cascade -cI PRM loop called autogenous regulation Uses of Recombinant DNA Technology, in general -Protein overproduction -Agriculture, drugs (insulin), chemical products -Vector with high-level production signals just before inserted DNA—overexpression of protein -Proper transcription and translation initiation signals -Gene Regulation -Hard to assay for particular gene -Would like to change conditions (i.e., add or subtract nutrients, chemicals, etc.) and see what happens -Expression vector—i.e., -galactosidase = reporter -Regulatory sequence of gene of interest attached to reporter gene -Re-insert into organism of choice and watch for any effects by assaying for reporter -DNA sequence analysis -DNA sequence leads to aa sequence which leads to protein structure -Can tell what the order of aa does when combine into certain order (distinct proteins) 3 -Tell DNA function from structure -Make mutations in test tube -Replace wild-type with mutant in organism -“Reverse Genetics” -Protein-DNA interactions -Quantify RNA synthesis -Use DNA (cloned gene) as probe for its mRNA product -Genome Projects -So far, 25 bacteria (most are known disease carriers), H. sapiens, D. melanogaster, C. elegans, S. cerevisiae -Evolution at molecular level Use of Recombinant DNA technology to Study Transcriptional Control -ALL Figures taken from Genetic Switch by Ptashne—Handout 2, 3 -DNAse I Footprinting—Protection Assay -Radio-label one end of DNA chain -Treat with DNAse I which cuts anywhere -Want Mild cut— ~1 cleavage per DNA molecule -Generate equivalent of DNA sequencing gel -But combined into 1 lane -Ladder with every step represented -If protein bound to DNA, then protect DNA from cleavage where protein bound -Will see gap in ladder = footprint -cI—OR interactions using DNAse I Footprinting Assay -At low cI concentration, see 2 sites with partial cleavage -OR1 and OR2 partially protected -At medium cI concentration, see 3 sites -OR1 and OR2 fully protected -OR3 partially protected -At high cI concentration, all 3 sites fully protected -When line up all 3 OL and OR, 2-fold symmetry almost exists -Table 2.1, 2.2 -12 out of 17 bases perfectly symmetrical -Consensus = TATCACCGC -1st A and C, 12/12 the same between all -3rd C, 11/12 the same between all -Down mutations at these 3 sites are strongest -cI—OR Binding Constant using DNAse I Footprinting Assay—Handout 3A -Relative amount of cI to get 50% protection -Cooperative = all 3 at same time -Intrinsic = each one individually operator cooperative intrinsic wt OR1 1 unit 2 units wt OR2 1 unit 25 units wt OR3 25 units 25 units -cI (dimer) at OR1 and OR2 help each other bind—Handout 3B 4 -Repressors at OR1 helped a little -Repressors at OR2 helped a lot -cI (dimer) at OR3 not interact with other cI operator cooperative intrinsic OR1C ----wt OR2 5 unit 25 units wt OR3 5 units 25 units -Knock out OR1, then OR2 and OR3 can interact—Handout 3C -OR2 binding is weaker since no interaction with OR1 (primary interactions) -OR3 binding is stronger since interactions now available -Genetic constructs used in reporter assay to show—Handout 2 Fig. 4.19 -On plasmid, -cI under control of lac operon promoter and operator -LacI gene also on plasmid -Construct 1 chromosome, -OR1-OR2-OR3-PRM-LacZ gene -No IPTG, then no cI, then PRM OFF -With IPTG, then cI, then PRM ON -Construct 2 chromosome, -OR3-OR2-OR1-PR-LacZ gene -No IPTG, then no cI, then PR ON -With IPTG, then cI, then PR OFF -When vary IPTG, leads to variation of active cI, leads to variation of -galactose synthesis -See effects of cI concentration on PR and PRM -Plot -gal levels as function of IPTG concentration -Active OR1, OR2, OR3—Handout 2 Table 4.2, 4.3 and figure on page -PR—-gal levels decreases as cI increases -PRM—-gal levels increase as OR1 and OR2 filled, and decreases as OR3 filled -Around same time that PR shut off, PRM turned on -Active OR1 with mutant OR2 and OR3—Handout 2 Fig. 4.21 top -PR—-gal levels decreases as cI increases -PRM—-gal levels never turned on -OR1 alone repress PR, not activate PRM -Active OR3 with mutant OR1 and OR2—Handout 2 Fig. 4.21 middle -PR—-gal levels never shut off -PRM—-gal levels never turned on -OR3 represses PRM, no effect on PR -Active OR2 with mutant OR1 and OR3—Handout 2 Fig. 4.21 bottom -PR—-gal levels decrease as cI increases -PRM—-gal levels increase as cI increases -OR2 alone activates PRM and represses PR -Need more repressor for each since no cooperative binding at OR1 and OR2 5 -Active OR2 and OR3 with mutant OR1—Handout 3D -PR—-gal levels decrease as cI increases -PRM—used PRM up-1 mutant–does not need cI to activate -Looks more like consensus --gal levels ON when no cI --gal levels decrease as cI increases -OR3 represses PRM -Summary Tables -xxx = cI dimer bound to site -ooo = mutated site so no binding of cI can occur lacZ—PRM OR3 OR2 OR1 PR—lacZ OFF ooo ooo xxx OFF ON ooo xxx ooo OFF OFF xxx ooo ooo ON OFF ---------ON ON ---xxx xxx OFF OFF xxx xxx xxx OFF -Last 3 are physiologically relevant - --- = empty, not mutated -Same experiments done with cro protein -cro bind also to OR1, OR2, OR3 -Tightly to OR3, then OR2, then OR1 -No cooperative action -Low cro, then shut off PRM, therefore shut off cI -High cro, then shut off PR 6