"Being Digital" or “Genetic Codes as Codes:”
advertisement

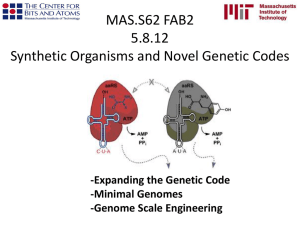
Towards a Theoretical Basis for Bioinformatics: “Genetic Codes as Codes” John R. Jungck Bioinformatics has developed primarily as a discipline within mathematics and computer science devoted to organizing and analyzing large biological databases. However, biology has much to offer to a synthetic discipline of bioinformatics that draws upon and respects the mutual contributions of biology, mathematics and computer science. In particular, biology has two major theoretical foundations, both evolutionary: namely, phylogenetic systematics and population genetics, that can serve as a cornerstone of a theoretical foundation of bioinformatics along with traditional empirically driven, pattern searching forms of classical bioinformatics. In this reconception of bioinformatics, mathematics and computer science are instrumental in developing biological theory and in solving practical biological problems. Since the genetic code is both an evolutionary product as well as a process for mediating the conversion of genotype to phenotype, it is argued here that an evolutionary analysis of genetic codes will fundamentally affect our ability to make meaning out of molecular messages through a theoretically grounded bioinformatics. Mathematical properties of genetic codes will be demonstrated with respect to their rates of transmission, correctability and detectability of errors, efficiencies, symmetries, and origins by employing coding theory (Baudot codes, Gray codes, Hamming codes, Huffman codes, common free codes, etc.), abstract algebra, graph theory, combinatorics, information theory, and phylogenetic systematics of sequences. Genetic codes become much more understandable and elegant to biologists, mathematicians, and computer scientists when they are not considered as mere ciphers, but are instead understood from three perspectives: codes per se, physical chemical interactions, and evolutionary selective pressures. These various faces of genetic codes are useful for making meaning out of molecular messages, applying causal mechanisms to complex patterns, and the efficient storage and retrieval of large complex data sets. In addition, some of the alternative distance metrics based upon different mathematical representations of genetic codes that have utility in genomic data base searching (comparative sequence analyses), phylogenetic tree construction, and prediction of three dimensional structure from primary structure will be illustrated and different evolutionary mechanisms affecting gene expression based upon codon usage will be considered. Key words: Evolutionary Bioinformatics; Genetic Codes; Huffman Codes (Fractals and Power Laws), Gray Codes, Hamming Codes, Baudot Codes, Comma-free Codes, Commaless Codes, and Overlapping Codes; codon usage; Gatlin-Grantham Hypotheses; Shannon’s Information Theory and Chaitin-Komogorov Algorithmic Complexity and Compressibility; Algebraic Coding Theory; Klein-4 groups. Bibliography: John R. Jungck, Ethel D. Stanley, and Marion Field Fass, Editors. (2002). Microbes Count! Problem Posing, Problem Solving, and Peer Persuasion in Microbiology. American Society for Microbiology Press: Washington, D.C. John R. Jungck, Editor, (1998-), The BioQUEST Library V & VI (2002). Academic Press: San Diego, California. John R. Jungck, (1998), Evolutionary Problem Solving. BioQUEST Notes 8 (2): 4-5 (February). John R. Jungck and Robert M. Friedman. 1984. Mathematical Tools for Molecular Genetics Data: An Annotated Bibliography. Bulletin of Mathematical Biology 46 (4): 699-744. John R. Jungck. 1984. The adaptationist programme in molecular evolution. The origins of genetic codes. In Molecular Evolution and Protobiology, K. Matsuno, K. Dose, K. Harada, and D. L. Rohlfing, eds., Plenum Press: New York, pp. 345-364. Martha O. Bertman and John R. Jungck. 1979. Group graph of the Genetic Code. Journal of Heredity 70: 379-384. John R. Jungck. 1978. The genetic code as a periodic table. Journal of Molecular Evolution 11: 211-224. Plus attach the PubMed bibliography on Codon Usage entitled: CodonCompPubMedBibliogr.doc Web site tools: 1. BioQUEST Curriculum Consortium 2. BEDROCK: Bioinformatics Education Dissemination: Reaching Out, Connecting, and Knitting-together 3. Biology Workbench 4. Codon Composition Analyzers: 5. 6. Freeland Lab: Genetic Code Evolution : projects a. A Bioinformatics Lab of the Biological Sciences Dept. at UMBC b. http://www.evolvingcode.net/project.php c. The CAI Calculator: measures codon usage bias in a gene d. Codon Sequence Analyzer: This tool investigates the codon error minimization property of the genetic code by analyzing protein coding sequences 7. Codon Usage Database (NAKAMURA Yasukazu, Dr.) a. http://www.kazusa.or.jp/codon/ 8. Codon Usage Table analysis i. http://www.entelechon.com/eng/cutanalysis.html ii. (Entelechon - the syntheticgenes.company) 9. Graphical Codon Usage Analyzer http://gcua.schoedl.de/ Markus Fuhrmann, Lars Ferbitz, Amparo Hausherr, Thomas Schödl and Peter Hegemann 10. The analysis of codon usage patterns. James O. McInerney < http://www.rfcgr.mrc.ac.uk/embnet.news/vol4_2/codon.html> 11.Correspondence Analysis of Codon Usage : CodonW is a programme designed to simplify the Multivariate analysis (correspondence analysis) of codon and amino acid usage. http://www.molbiol.ox.ac.uk/cu/ 12.