genetic transformation - Division of Teaching Labs
advertisement

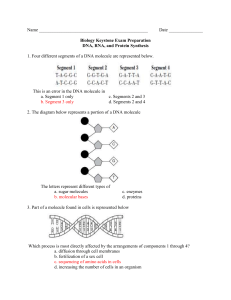
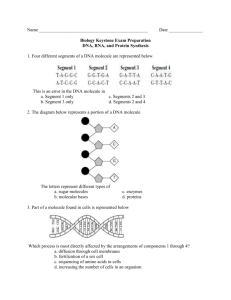
GENETIC TRANSFORMATION: SEQUENCE EXPLORATION You will use a DNA search engine called Nucleotide BLAST at the National Center for Biotechnology Information (NCBI) website to search GenBank, a fully annotated database of all publicly available DNA sequences and their protein translations, for the foreign gene used in the Genetic Transformation Lab. Sequences in GenBank are contributed by individual labs and sequencing facilities all over the world. As of April 2008, there were more than 76 million individual sequences from over 260,000 organisms in the GenBank database. PART I: BLAST N SEARCH Below is the wild-type (natural, unaltered) DNA sequence of the foreign gene which we have been exploring in our Genetic Transformation Lab. This is GeneG that is contained in the pYSPG plasmid that some of you transformed into bacteria. This gene is 717 nucleotide bases long, including the start codon (ATG) and the stop codon (TAA). Here is the sequence of GeneG: >>GeneG 1 61 121 181 241 301 361 421 481 541 601 661 ATGAGTAAAG GATGTTAATG AAACTTACCC GTCACTACTT CATGACTTTT AAAGATGACG AATAGAATCG ATGGAATACA ATCAAAGTTA CATTATCAAC CTGTCCACAC CTTGAGTTTG GAGAAGAACT GGCAAAAATT TTAATTTTAT TCTCTTATGG TCAAGAGTGC GGAACTACAA AGTTAAAAGG ACTATAACTC ACTTCAAAAT AAAATACTCC AATCTGCCCT TAACAGCTGC TTTCACTGGA CTCTGTCAGT TTGCACTACT TGTTCAATGC CATGCCCGAA GACACGTGCT TATTGATTTT ACATAATGTA TAGACACAAC AATTGGCGAT TTCCAAAGAT TAGGATTACA GTGGTCCCAG GGAGAGGGTG GGGAAGCTAC TTCTCAAGAT GGTTATGTAC GAAGTCAAGT AAAGAAGATG TACATCATGG ATTAAAGATG GGCCCTGTCC CCCAACGAAA CATGGCATGG TTCTTGTTGA AAGGTGATGC CTGTTCCATG ACCCAGATCA AGGAAAGAAC TTGAAGGTGA GAAACATTCT GAGACAAACC GAAGCGTTCA TTTTACCAGA AGAGAGATCA ATGAACTATA ATTAGATGGC AACATACGGA GCCAACACTT TATGAAACAG TATATTTTAC TACCCTTGTT TGGACACAAA AAAGAATGGC ATTAGCAGAC CAACCATTAC CATGATCCTT CAAATAA 1. To access the NCBI Nucleotide BLAST webpage, right-click on the following link and select ‘Open Hyperlink’: http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS= megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=bla sthome 2. Where it says ‘Enter Query Sequence’, copy and paste the GeneG sequence (including the >>GeneG header) into the box provided. It does not matter if there are numbers in the sequence. (This is illustrated on the following page.) 1 3. Where it says ‘Choose Search Set’, select ‘Nucleotide Database (nr/nt)’ from the drop-down Database menu. 4. Leave all other fields blank. 5. Click on the button at the bottom left side of the page. 6. Wait a few moments while the program searches GenBank for a match to the foreign gene. 7. You should now have a page with your search results. Scroll-down to the ‘Descriptions’ section and look at the results. Notice that they are ranked by scores, 2 from highest to lowest. Score indicates the degree of identity between your query sequence and the sequence in the database. Accession Number is the identifier that has been assigned to a particular sequence in the database. 8. Click on the ‘Accession Number’ link for the top hit, i.e. the one with the highest score which should be at the top of the list. This will take you to the GenBank information page for that sequence. What protein is encoded by GeneG? What organism is this gene from? PART II: DNA SEQUENCE ALIGNMENT The pYSPB plasmid that some of you transformed into bacteria contains a mutated version of the foreign gene, designated GeneB. Below is the sequence of GeneB: >>GeneB 1 61 121 181 241 301 361 421 481 541 601 661 ATGAGTAAAG GATGTTAATG AAACTTACCC GTCACTACTT CATGACTTTT AAAGATGACG AATAGAATCG ATGGAATACA ATCAAAGTTA CATTATCAAC CTGTCCACAC CTTGAGTTTG GAGAAGAACT GGCAAAAATT TTAATTTTAT TCTCTCACGG TCAAGAGTGC GGAACTACAA AGTTAAAAGG ACTATAACTC ACTTCAAAAT AAAATACTCC AATCTGCCCT TAACAGCTGC TTTCACTGGA CTCTGTCAGT TTGCACTACT TGTTCAATGC CATGCCCGAA GACACGTGCT TATTGATTTT ACATAATGTA TAGACACAAC AATTGGCGAT TTCCAAAGAT TAGGATTACA GTGGTCCCAG GGAGAGGGTG GGGAAGCTAC TTCTCAAGAT GGTTATGTAC GAAGTCAAGT AAAGAAGATG TACATCATGG ATTAAAGATG GGCCCTGTCC CCCAACGAAA CATGGCATGG TTCTTGTTGA AAGGTGATGC CTGTTCCATG ACCCAGATCA AGGAAAGAAC TTGAAGGTGA GAAACATTCT GAGACAAACC GAAGCGTTCA TTTTACCAGA AGAGAGATCA ATGAACTATA ATTAGATGGC AACATACGGA GCCAACACTT TATGAAACAG TATATTTTAC TACCCTTGTT TGGACACAAA AAAGAATGGC ATTAGCAGAC CAACCATTAC CATGATCCTT CAAATAA You will use an online program to compare the sequences of the wild-type (GeneG) and mutated (GeneB) genes; this is known as a DNA sequence alignment. An alignment uses an algorithm (a step-by-step procedure) to compare the order of nucleotide bases in the sequences and then lines them up so that the number of identical bases is maximized. The alignment program will point out those bases that are identical (indicated by an asterisk - ), those that are similar (:), and those that are completely different (no symbol). Alignments are useful to study how closely genes are related which then allows evolutionary relationships to be determined. For example, how are genes that code for the same protein in different species or genes passed on between generations related? You will use the ClustalW2 general sequence alignment tool provided by the European Bioinformatics Institute. 3 1. Right-click on the following link and select ‘Open Hyperlink’ to access the ClustalW2 website: http://www.ebi.ac.uk/Tools/clustalw2/index.html 2. Where it says “Enter or paste a set of sequences in any supported format” you will paste the GeneB and GeneG sequences. a. First copy and paste the GeneG sequence (including the >>GeneG header) into the box. b. Press Enter to leave a space after the GeneG sequence. c. Then copy and paste the GeneB sequence (including the >>GeneB header) below the GeneG sequence in the same box. 3. There is no need to change any of the default parameters or fill in any other fields on the page. 4. Click on 5. You may have to wait a few moments while the alignment program is running. 4 6. Scroll-down the results page to the “Alignment” section. Click on the “View Alignment File” button. Copy and paste the alignment results into the box provided below. Can you identify which nucleotide bases have been mutated in GeneB? Alignment of GeneG and GeneB Nucleotide Sequences: 5 PART III: DNA TRANSLATION AND PROTEIN SEQUENCE ALIGNMENT The ClustalW2 program can also be used to align protein (amino acid) sequences. First, you will use a tool provided by Colorado State University to translate the sequences of GeneG and GeneB into their corresponding amino acid sequences. Then, you will use the ClustalW2 program to align the amino acid sequences and identify which amino acid(s) is mutated in GeneB. 1. Right-click on the following link and select ‘Open Hyperlink’ to access the website: http://www.vivo.colostate.edu/molkit/translate/ 2. In the first empty box, copy and paste the sequence for GeneG – this time, DO NOT include the >>GeneG header (but numbers can be included). 3. Click on ‘Translate DNA’. (Do not change any of the other options.) 4. Click on ‘Text Output’. 5. You should see the amino acid translation above the DNA sequence. Amino acids are given in one-letter code. Recall that three nucleotide bases code for one amino acid. 6. In the third drop-down menu from the left it currently says ‘Amino acids and DNA’, select ‘Amino acids only’. 7. You should now see only the amino acid sequence translated from the gene. 6 8. Copy and paste the amino acid translation for GeneG (from the number 1 to the end of the sequence) in the space provided below. 9. Repeat steps 2 to 8 for GeneB. >>GeneG_aminoacid >>GeneB_aminoacid 7 10. Go back to the ClustalW2 website (see pg.4 instructions). This time, copy and paste the amino acid sequences translated from GeneG and GeneB into the query box (one below the other, including the >> headers). 11. Click on ‘Run’. 12. Below, you are provided with the one- and three-letter abbreviations for the 20 common amino acids. Examine the alignment results. Can you identify what amino acid mutation has occurred in GeneB? One letter code A C D E F G H I K L M N P Q R S T V W Y Three letter code ala cys asp glu phe gly his ile lys leu met asn pro gln arg ser thr val try tyr Amino acid alanine cysteine aspartic acid glutamic acid phenylalanine glycine histidine isoleucine lysine leucine methionine asparagine proline glutamine arginine serine threonine valine tryptophan tyrosine 13. Can you manually transcribe a DNA sequence into its complementary RNA and translate that RNA into its corresponding amino acid sequence? Of course! Scientists have uncovered the Universal Genetic Code which describes how a specific three nucleotide codon translates for a particular amino acid. Right-click on the following link and select ‘Open Hyperlink’: http://learn.genetics.utah.edu/content/begin/dna/transcribe/ 14. After you have gone through the exercise, go back to your nucleotide sequence alignment results (on pg. 5). Can you identify the codon responsible for the amino acid mutation seen in GeneB? 8 When you go back to the lab later today, you will discover what effect this mutation has on the phenotype of the E.coli cells which have been transformed with pYSPB compared to pYSPG! PART IV: DISCOVER MORE! Use your favourite search engine (e.g. Google) to discover more about the foreign gene (GeneG) and the protein it encodes! What are some special properties of this protein? Who won the Nobel Prize for its discovery and development? What are some of the ways scientists have used this gene for scientific research? WANT TO LEARN MORE ABOUT DNA SCIENCE?! DNA Today! To find out more about the role of DNA science in your lives, join commentators Dave Micklos and Jan Witkowski from world-renowned Cold Spring Harbor Laboratory for a lively discussion of DNA in the news: http://www.dnalc.org/ddnalc/dna_today/index.html DNA from the Beginning! Visit http://www.dnaftb.org/ to discover the concepts and experiments that define the fields of genetics and molecular biology. This animated primer features the work of over 100 scientists and researchers. DNA Timeline! Travel through time with scientists from the first discovery of DNA to sequencing of the human genome at http://www.dnai.org/timeline/index.html. To learn more about GenBank, visit: http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18073190 Note: For the purposes of these exercises the wild-type gene sequence of GFP was provided as GeneG, in actuality the gene transformed into bacteria in the laboratory experiment was EGFP (Enhanced GFP) which contains several amino acid substitutions which allow the fluorescence of the protein to be brighter and last longer. The sequence for GeneB given in this exercise was constructed based on the knowledge that a single amino acid point mutation (Y66H) is responsible for the blue fluorescence seen in the GFP variant known as BFP (Heim et al., PNAS USA, 91:12501-12504, 1994); in actuality the gene transformed into bacteria in the laboratory exercise was EBFP (Enhanced BFP) which in addition to the Y66H mutation, contains several other amino acid mutations which similarly allow the blue fluorescence to shine brighter and last longer. 9