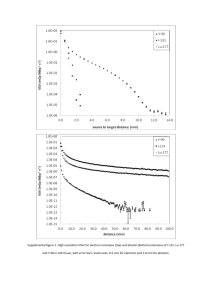
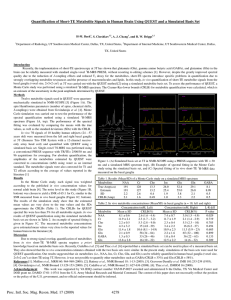
Supplementary Methods: 1H-MRS data acquisition and analysis 1H
advertisement

Supplementary Methods: 1H-MRS data acquisition and analysis 1H-MRS acquisition 1H-MRS spectra were acquired on a General Electric (Milwaukee, Wisconsin, USA) 3 Tesla HDx MR system. As detailed previously,1, 2 subjects underwent an initial localizer scan followed by acquisition of structural images including an axial 2D T2weighted fast spin echo scan and an axial fast fluid-attenuated inversion recovery scan (total scan time = 5 min). These were followed by a whole-brain threedimensional coronal inversion recovery prepared spoiled gradient echo (IR-SPGR) scan, giving isotropic 1.1-mm voxels in a scan time of approximately 6 min (echo time (TE) = 2.82 msec; repetition time (TR) = 6.96 msec; inversion time = 450 msec; excitation flip angle = 20°). 1H-MRS spectra (PRESS—Point RESolved Spectroscopy; TE = 30 msec; TR = 3000 msec; 96 averages) were acquired using the standard GE PROBE (PROton Brain Examination) sequence, which uses a standardized CHEemical Shift Selective (CHESS) water suppression routine, as previously described.1, 2 For each metabolite spectrum, unsuppressed water reference spectra (16 averages) were also collected as part of the acquisition. Shimming was optimized, with auto-prescan performed twice before each scan. Using standardized protocols, voxels were prescribed in the left thalamus (15 x 20 x 20 mm right–left, anterior–posterior, superior–inferior) and anterior cingulate cortex (ACC, 20 x 20 x 20 mm). The left thalamic voxel was positioned to include the maximum amount of grey matter based on individual anatomy. The ACC voxel was prescribed from the midline sagittal localizer, with the center of the voxel placed 13 mm above the genu of corpus callosum perpendicular to the AC–PC line. 1H-MRS quantification For compatibility with our previous report1 spectra were analyzed using LCModel version 6.1-4 F3, using a standard basis set of 16 metabolites (L-alanine, aspartate, creatine, phosphocreatine, ϒ-amino butyric acid (GABA), glucose, glutamine, glutamate, glycerophosphocholine, glycine, myo-inositol, L-lactate, Nacetylaspartate (NAA), N-acetylaspartylglutamate, phosphocholine, and taurine), acquired with the same field strength (3 Tesla), localization sequence (PRESS) and echo time (30 msec). Model metabolites and concentrations used in the basis set are fully detailed in the LCModel manual (http://s-provencher-.com/pages/lcmmanual.shtml). Poorly fitted metabolite peaks (Cramer–Rao minimum variance bounds (CRLB) of >20% as reported by LCModel) were excluded from further analysis. NAA is reported in combination with N-acetylaspartylglutamine (NAAG). For all metabolites present in the 1H-MRS spectra, quantitative concentrations were calculated in units of mmol, but to indicate that these values have not been scaled to allow for the effects of T1 or T2 relaxation, results are presented as ‘institutional units’. The IR-SPGR scans were used for localization of the spectroscopy voxels and were segmented into grey matter, white matter and cerebrospinal fluid (CSF) using Statistical Parametric Mapping 2 software (SPM2; Wellcome Trust Centre for Neuroimaging, Institute of Neurology, University College London, United Kingdom) to allow correction of water-scaled 1H-MRS metabolite values for partial volume CSF contamination. Metabolite values were corrected for the presence of CSF in the voxel using the formula Mcorr = M/(wm +gm), where M is the uncorrected metabolite value, and wm and gm are the white and grey matter fractions from the segmented images (ranging between 0 and 1) for the spectroscopy voxel. Since the metabolite measures were derived with water-scaling, a further correction was applied to correct the estimated water concentration of the voxel for partial volume CSF contamination, assuming a CSF water concentration of 55,556 mol/m3 and the LCModel default brain water concentration of 35,880 mol/m3 (http://s-provencher.com/pages/lcm-manual.shtml). For practical purposes, these two correction factors were combined into a single equation which simplifies to Mcorr = M*(wm+gm + 1.55CSF) / (wm+gm), where CSF is the fraction of the voxel filled with cerebrospinal fluid. 1. 2. 3. Stone JM, Day F, Tsagaraki H, Valli I, McLean MA, Lythgoe DJ, O'Gorman RL, Barker GJ, McGuire PK. Glutamate dysfunction in people with prodromal symptoms of psychosis: relationship to gray matter volume. Biol.Psychiatry. 2009;66(6):533-539. Egerton A, Brugger S, Raffin M, Barker GJ, Lythgoe DJ, McGuire PK, Stone JM. Anterior cingulate glutamate levels related to clinical status following treatment in first-episode schizophrenia. Neuropsychopharmacology. 2012;37(11):2515-2521. Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson.Med. 1993;30(6):672-679.