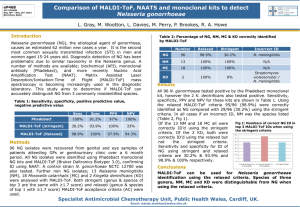
Enhancing the cellular uptake of Py–Im polyamides through next

Supporting Information
Enhancing the cellular uptake of Py-Im polyamides through next-generation aryl turns
Jordan L. Meier, David C. Montgomery, and Peter B. Dervan*
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125
Table of Contents for Supporting Information
Page
Supplemental Figures
Primer sequences for qRT-PCR
Sample calibration curve for quantitative flow cytometry experiments
Reagents and Equipment
General synthetic procedure for conjugation of
-aryl turn
S2
S8
S9
S10
S10
Polyamide characterization data
Synthetic schemes
References
S11
S12
S15
S1
Figure S1. Inhibition of nuclear-receptor mediated gene expression by
-aryl polyamides. (a) Effect of
turn polyamides 2 and 4 on dexamethasone (DEX)-induced FKBP5 gene expression as measured by qRT-
PCR analysis. I = DEX-induced. NI = non-induced. Polyamide 2 concentrations: 100, 1000, 10,000 nM, polyamide 4 concentrations: 1, 10, 100 nM. (b) Effect of
-turn polyamides 2 and 4 on dihydroxytestosterone (DHT)-induced FKBP5 gene expression as measured by qRT-PCR analysis.
Polyamide 2 concentrations: 100, 1000, 10,000 nM, polyamide 4 concentrations: 1, 10, 100 nM. I = induced. NI = not induced.
S2
Figure S2. Time courses for (a) cytotoxicity analysis, (b) dexamethasone-induced gene expression studies, (c) dihydroxytestosterone-induced gene expression studies, (d) flow cytometric analysis of uptake of polyamide-FITC conjugates , (e) confocal microscopy analysis of uptake of polyamide-FITC conjugates.
S3
Figure S3. Relative cytotoxicity of fluorescent analogues 13 and 14 in A549 cells. Cytotoxicity of the parent compounds 2 and 4 are found in Figure 2 of the main text.
Figure S4.
Percentage of cells labeled as a function of time exposed to
-turn polyamides
-aryl 13 20
(100 nM). (a) Bar graph depicting relative percentage of cells fluorescently labeled by
-amino polyamides
13 and 17 compared to their
-aryl analogues 14 and 18 . (b) Complete data for all polyamide cores analyzed in this study. Percentage of cells labeled is calculated relative to control cells treated with vehicle
S4
DMSO.
S5
Figure S5. Analysis of
-aryl ( 14 ) and
-amino ( 13 ) polyamide efflux. (a) Adherent A549 cells were incubated with 100 nM 13 or 14 in the presence or absence of verapamil (10
M) for 48 hrs and analyzed by flow cytometry. Both polyamides show similar increases in uptake. All samples were analyzed in the same biological experiment. Cellular fluorescence reported as arbitrary units. (a) Adherent A549 cells were incubated with 1000 nM 13 or 14 in the presence or absence of verapamil (10
M) for 24 hrs and analyzed by flow cytometry. Both polyamides show similar increases in uptake. (c) Washout study of polyamides.
A549 cells were treated with 13 or 14 for 24 hrs, at which point polyamides were removed and cells were
S6
grown in fresh media for 0, 24, or 48 hr followed by FACS analysis. Both compounds show similar profiles, suggesting efflux or dilution by cell growth proceeds similarly for each.
Figure S6. Confocal microscopy analysis of
-aryl polyamide nuclear localization. Adherent A549 cells were treated with 1 μM
-aryl polyamide-FITC conjugates ( 14 / 16 / 18 / 20 ) for 16 hrs, and counterstained with
S7
Hoechst 33342 (nuclear stain) and Mitotracker Red CM-H
2
Ros (mitochondrial stain) just prior to imaging.
Top left: polyamide-FITC (
ex
= 488 nm
em
= 505-530 nm). Top right: Hoechst (
ex
= 750 nm [2-photon]
em
= 390-465 nm). Bottom left: Mitotracker (
ex
= 543 nm
em
= 565-615 nm). Bottom right: three-color overlay.
Figure S7. Cytotoxicity of
-aryl polyamide cores conjugated to a C-terminal isophthalic acid (IPA) tails in
A549 cells. Complete structures are given in Scheme S4.
Figure S8. Inhibition of hypoxia-induced gene expression by
turn polyamides targeted to the hypoxia response element
(HRE) of the vascular endothelial growth factor ( VEGF) locus.
The effect of
-substituted polyamides 21 and 22 on desferoxamine (DFO)-induced VEGF gene expression was analyzed by qRT-PCR analysis. I = DFO induced. NI = noninduced. Polyamide 21 conenctrations: 100, 1000, 10,000 nM.
Polyamide 22 concentrations: 10, 100, 1000 nM.
S8
S9
Primer Sequences for qRT-PCR Experiments
GUSB:
Fwd : 5’-CTCATTTGGAATTTTGCCGATT-3’
Rev : 5’-CCGAGTGAAGATCCCCTTTTTA-3’
GILZ:
Fwd : 5’-TGGTGGCCATAGACAACAAG-3’
Rev : 5’-TGCTCCTTCAGGATCTCCAC-3’
FKBP5:
Fwd : 5’-CGGAAAGGAGAGGGATATTCA-3’
Rev : 5’-CCACATCTCTGCAGTCAAACA-3’
KLK3 (PSA):
Fwd : 5’-TCTGCGGCGGTGTTCTG-3’
Rev : 5’-GCCGACCCAGCAAGATCA-3’
VEGF:
Fwd : 5’-AGGGCAGAATCATCACGAAG-3’
Rev : 5’-GGGTACTCCTGGAAGATGTCC-3’
S10
Representative Calibration Curve for Flow Cytometry Experiments
Sample calibration curve for Sphero Rainbow Calibration Particles used to calculate molecules of equivalent fluorescence (MEFL). Calculation of MEFL for polyamide-treated cells was used to estimate the cellular polyamide concentration (see main text).
S11
Reagents and Equipment
Anhydrous N , N -dimethylformamide (DMF), diisopropylethylamine (DIEA), triethylsilane, trifluoroacetic acid
(CF
3
CO
2
H [peptide synthesis grade]), isophthalic acid, and (±)-verapamil hydrochloride were purchased from Sigma-Aldrich. Fluorescein isothiocyanate (FITC, Isomer I) was purchased from Invitrogen. Kaiser oxime resin (LL, 200-400 mesh) and benzotriazole-1-yl-oxy-trispyrrolidinophosphonium hexafluorophosphate (PyBOP) were from Novabiochem. Nβ-Cbz-N-γ-Boc-D-3,4-diaminobutyric acid [Z-D-
β-Dab(Boc)-OH] was purchased from Sigma Aldrich (product code 28206). All Boc-protected N methylpyrrole and Nmethylimidazole monomers and dimers for polyamide synthesis were prepared according to the published protocols (51,52). Bulk grade solvents were purchased fom Fisher Scientific.
Centrifugation was performed in a Beckman Coulter bench-top centrifuge (Allegra 21R) equipped with a
Beckman swing-out rotor (model S4180). Preparative HPLC purification was performed on an Agilent 1200
Series instrument equipped with a Phenomenex Gemini preparative column (250 x 21.2 mm, 5 μm) with the mobile phase consisting of a gradient of acetonitrile (MeCN) in 0.1% CF
3
CO
2
H (aqueous). Polyamide synthesis was monitored by analytical HPLC, with analysis conducted on a Beckman Gold instrument equipped with a Phenomenex Gemini analytical col umn (250 x 4.6 mm, 5μm), a diode array detector, and the mobile phase consisting of a gradient of MeCN in 0.1% CF
3
CO
2
H (aqueous). Polyamide concentrations were measured by UV analysis on a Hewlett-Packard model 8453 diode array spectrophotometer in distilled and deionized water (ddH
2
O) with a molar extinct ion coefficient (ε) of 69,500 M -1 cm -1 at λ max
of 310 nm.
General synthetic procedure for conjugation of
-aryl turn
A solution of benzoic acid (2.4 mg, 0.02 mmol) and PyBOP (10.4 mg, 0.02 mmol) in DIEA (14 μL, 0.2 mmol) and DMF (0.4 mL) was stirred at 23 °C for 1 hour. Separately, polyamide 2 (800 nmol) was dissolved in DMF (0.1 mL) and slowly added to the pre-activated isophthalic acid solution. The reaction was allowed to stand at 23 °C for 1 hr and monitored by analytical HPLC. Upon completion, Et
2
O (4.5 mL) was added and the reaction was vortexed thoroughly, resulting in formation of a brownish white precipitate that was isolated by centrifugation (~ 4500 rpm) (21). After removal of the organic layer, the residual solid was dissolved in DMF (0.5 mL) with ultrasonication, and diluted with 20% MeCN in 0.1% CF
3
COOH (5 mL).
Purification by reverse-phase HPLC followed by lyophilization provided
-aryl polyamide 4 (0.36 mmol,
45%). MS (MALDITOF) cald’d for C
72
H
81
N
22
O
13
[M+H] + 1461.6, found 1461.9.
S12
Polyamide characterization data
1 MS (MALDITOF) cald’d for C
65
H
76
N
21
O
12
[M+H] + 1342.6, found 1342.9.
2 MS (MALDITOF) cald’d for C
65
H
77
N
22
O
12
[M+H] + 1357.6, found 1357.8.
3 MS (MALDITOF) cald’d for C
67
H
79
N
22
O
13
[M+H] + 1399.6, found 1399.9.
4 MS (MALDI-
TOF) cald’d for C
72
H
81
N
22
O
12
[M+H] + 1461.6, found 1461.9.
5 MS (MALDITOF) cald’d for C
71
H
80
N
23
O
13
[M+H] + 1462.6, found 1462.7.
6 MS (MALDITOF) cald’d for C
72
H
80
N
23
O
15
[M+H] + 1506.6, found 1506.5.
7 MS (MALDITOF) cald’d for C
72
H
82
N
23
O
13
[M+H] + 1476.6, found 1476.2.
8 MS (MALDITOF) cald’d for C
78
H
85
N
22
O
13
[M+H] + 1537.7, found 1537.5.
9 MS (MALDITOF) cald’d for C
73
H
81
N
22
O
15
[M+H] + 1505.6, found 1505.8.
10 MS (MALDITOF) cald’d for C
73
H
81
N
22
O
15
[M+H] + 1505.6, found 1505.8.
11 MS (MALDITOF) cald’d for C
78
H
93
N
24
O
14
[M+H] + 1589.7, found 1589.8.
12 MS (MALDITOF) cald’d for C
78
H
93
N
24
O
14
[M+H] + 1589.7, found 1589.8.
13 MS (MALDITOF) cald’d for C
78
H
83
N
23
O
14
S [M+H] + 1597.6, found 1598.0.
14 MS (MALDITOF) cald’d for C
85
H
87
N
23
O
15
S [M+H] + 1701.6, found 1701.8.
15 MS (MALDITOF) cald’d for C
77
H
80
N
22
O
14
S
2
Cl
1
[M+H] + 1635.5, found 1635.6.
16 MS (MALDITOF) cald’d for C
84
H
83
N
22
O
15
S
2
Cl
1
[M+H] + 1738.6, found 1738.3.
17 MS (MALDITOF) cald’d for C
77
H
80
N
24
O
14
S
1
[M+H] + 1596.6, found 1596.8.
18 MS (MALDITOF) cald’d for C
84
H
86
N
24
O
15
S
1
[M+H] + 1702.6, found 1702.8.
19 MS (MALDI-
TOF) cald’d for C
70
H
80
N
23
O
14
S
1
[M+H] + 1498.6, found 1498.9.
20 MS (MALDITOF) cald’d for C
77
H
83
N
23
O
15
S
1
[M+H] + 1601.6, found 1601.7.
21 MS (MALDITOF) cald’d for C
64
H
73
N
21
O
12
S
1
Cl
1
[M+H] + 1394.5, found 1394.6.
22 MS (MALDI-TOF) cald’d for C
71
H
76
N
21
O
13
S
1
Cl
1
[M+H] + 1497.5, found 1497.7.
23 MS (MALDI-
TOF) cald’d for C
64
H
74
N
23
O
12
[M+H] + 1356.6, found 1356.8.
24 MS (MALDITOF) cald’d for C
71
H
77
N
23
O
13
[M+H] + 1459.6, found 1459.6.
25 MS (MALDITOF) cald’d for C
57
H
73
N
22
O
12
[M+H] + 1257.6, found 1257.5.
26 MS (MALDITOF) cald’d for C
64
H
76
N
22
O
13
[M+H] + 1360.6, found 1360.8.
S13
Scheme S1. Monomers, dimers, and general scheme for solid-phase synthesis of polyamides analyzed in this study.
S14
Scheme S2. Scheme for solution-phase derivativization of polyamides and conjugation of
-aryl turns.
S15
Scheme S3. Scheme and complete chemical structures for fluorescent polyamides 14 20 .
S16
Scheme S4. Complete structures of polyamides 21 26 .
S17
References:
51
. Baird, E.E. and Dervan, P.B. (1996) Solid Phase Synthesis of Polyamides Containing Imidazole and
Pyrrole Amino Acids. J Am Chem Soc ,
118
, 6141-6146.
52. Chenoweth, D.M., Harki, D.A. and Dervan, P.B. (2009) Solution-phase synthesis of pyrroleimidazole polyamides. J Am Chem Soc ,
131