Alleles at the Blp locus in barley determine if the
advertisement

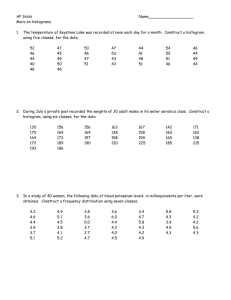
Homework # 2 Due January 28 1. Alleles at the Blp locus in barley determine if the mature inflorescence (the “head”) is black or white. See http://barleyworld.org/oregonwolfe/images for pictures of the Oregon Wolfe Barley (OWB) populations. For this assignment, choose the n = 82 Hordeum bulbosumderived doubled haploid population. Note: The OWB DH progeny are not numbered consecutively, but there are 82 of them. A. Draw (or use Excel to make – see instruction at the INFO link for the Mendelian genetics section) a frequency distribution showing the number of BlpBlp (black-headed) and blpblp (white-headed) doubled haploid progeny. Some heads may appear to be intermediate in color. For this homework, however, every head has to be described as black or white. In your histogram, be sure to indicate the phenotype of each of the parents. B. Which term best describes this frequency distribution? a. Discontinuous b. Continuous C. How many loci do you hypothesize are responsible for determining head color in this population? D. Calculate a chi square to test your hypothesis regarding the number of loci that determine head color in this population – fill in the following table. Note: Parents are NOT used for calculating the chi square test. Phenotype # Observed (O) # Expected (E) Totals 82 82 O-E E. Calculated X2 = _______ ; df = _______ ; probability = _______ F. Do you accept or reject your hypothesis? G. How many alleles are segregating at the locus (or each of the loci) that you have determined are responsible for head color? (O-E)2/E 2. Histograms can assist in developing preliminary hypotheses regarding the number of genes determining a particular phenotype. In question # 1, you created a histogram for the “head color” trait in barley. As progressively more genes determine a phenotype, the histograms will shift from discontinuous (indicating qualitative inheritance, few genes) to continuous (indicating quantitative inheritance, multiple genes). To demonstrate this to your own satisfaction, graph histograms for the following scenarios. In all cases assume that you have derived 100 doubled haploids from an F1 plant obtained by crossing two completely inbred parents. A. Assume the trait is determined by alleles at 2 loci. a. The cross is AAbb x aaBB b. Assume each dominant allele at each locus is worth 2 “units” and each recessive allele is worth 1 “unit”. c. Determine the value of each of the 4 possible doubled haploid classes and make a histogram where the X axis is “units of worth” and the Y axis is number of lines in each class of “units of worth”. d. Be sure to show the values of the parents on the histogram. B. Assume the trait is determined by alleles at 4 loci. a. The cross is AAbbCCdd x aaBBccDD. b. Assume each dominant allele at each locus is worth 2 “units” and each recessive allele is worth 1 “unit”. c. Determine the value of each of the 16 possible doubled haploid classes and make a histogram where the X axis is “units of worth” and the Y axis is number of lines in each class of “units of worth”. d. Be sure to show the values of the parents on the histogram.