First demonstration on NSE biomarker of a computational
advertisement

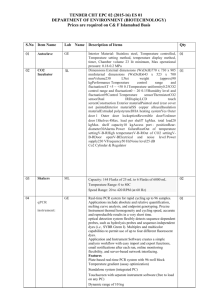
First demonstration on NSE biomarker of a computational environment dedicated to lab-on-chip based cancer diagnosis Pierre GRANGEAT1, Laurent GERFAULT1, Caroline PAULUS1, Vangelis KRITSOTAKIS2, Manolis TSIKNAKIS2, Frédérique LISACEK3, Pierre-Alain BINZ4, Manuel M PEREZ-PEREZ5, Mathieu TRAUCHESSEC6, Virginie BRUN6. CEA, LETI, MINATEC, DTBS, Laboratoire d’Electronique et de Systèmes pour la Santé, 17 rue des Martyrs, F-38054 Grenoble cedex 9, France. 2 FORTH, Foundation for Research and Technology –ICS, Vassilika Vouton, Heraklion 71110, Greece 3 Proteome Informatics Group, Swiss Institute of Bioinformatics, Michel-Servet 1, CH-1211 Geneva 4, Switzerland 4 Geneva Bioinformatics (GeneBio) SA, Av de Champel 25, CH-1206 Geneva, Switzerland 5 ATOS Research and Innovation, Biotechnology and Health Unit,C/ Albarracin 25, Madrid 28035, Spain 6 CEA/INSERM/UJF U880, Laboratoire d’Etude de la Dynamique des Protéomes, 17 rue des Martyrs, F-38054 Grenoble cedex 9, France. 1 Introduction : CEA-LETI is designing new silicon-based digestion and nano-chromatography components for mass spectrometry analytical chains. EDyP laboratory has developed an expertise dedicated to the absolute and accurate quantification of proteins in biofluids [1]. On the LOCCANDIA European project (www.loccandia.eu) an innovative integrated clinicoproteomics computational environment has been developed to support the LOCCANDIA integrated lab-on-chip based diagnostic device [2]. It combines several modules dedicated to web-based data and parameter management, profile reconstruction for protein quantification, data visualisation, peptide identification, and decision support based on classification. In this communication, we propose the first demonstration of this software environment on an experimental data set. Method : This LIMS computational environment integrates several innovative contributions. It is a web-based application, responsible for the storage, examination and manipulation of clinical and proteomic data. The three-tier architecture allows any upgrade as requirements or technology change. The profile reconstruction module computes the protein quantification from LC-MS spectrograms. The new version implements the inversion of a nano-liquid chromatography dynamical model [3] that allows including non-linear isotherms as it is the case for nano-LC chip. The visualization module is the MSight software platform. It provides a sophisticated way of visualizing mass spectrometer raw data. The identification module from LC/MS/MS data is the Phenyx software[4]. The classification module is based on Logistic regression and Support Vector Machines with nine different kernels. Preliminary data : Neuron Specific Enolase (NSE), recognized as a lung and brain cancer biomarker, was chosen as model. Using the PSAQ (Protein Standard Absolute Quantification) strategy [5], EDyP laboratory has developed a LC-MS-based assay enabling the absolute quantification of NSE in serum samples. The analytical process includes an immunoenrichment step followed by trypsin digestion and LC-MS analysis on an Orbitrap mass spectrometer. Quantification is performed by spiking into the samples an isotopically-labeled version of NSE as internal standard. To generate the data set, 4 defined amounts of unlabeled NSE (ranging from 0 to 200 ng/mL) were artificially spiked into 1 mL serum samples. NSE was accurately quantified down to its physiological level (50ng/mL). The data have been uploaded into the LIMS via the web-based access and the relevant parameters have been filled. The analysis has been carried on 3 peptides (IEEELGDEAR, GNPTVEVDLYTAK, IVIGMDVAASEFYR). The identification module has identified these peptides and transferred the information to the visualisation module for annotation purposes. The visualisation module clearly demonstrates the presence of these peaks in the light and heavy forms. The reconstruction module has been applied independently on each peptide. Calibration is performed to estimate the common model parameters. Regions of interest around processed peptides are specified. Then, unsupervised adaptive computation of protein concentration is performed. For the 27 computed protein quantities, the maximum relative error is 20% with a mean value of 10%. Despite 3% of retention time variation and 35% of gain variation, the mean coefficient of variation on quantification is 10%. [1] Brun et al. (2009), J. Proteomics 72.5, 740-749 [2] Jordan et al., (2008), 5th pHealth Workshop, Valencia, Spain. [3] Paulus et al. (2009), SYSID 2009, 251-256, Saint Malo, France. [4] Colinge et al. (2004) Proteomics 7, 1977-84. [5] Brun et al. (2007), Mol. and Cell. Proteomics 6.12, 2139-2149. Novel aspect: Protein quantification computational environment, web-based application, nano liquid chromatography dynamical model, unsupervised adaptive computation, NSE quantification in serum samples.