CW7

Tools in Bioinformatics
Class work 7
Please pay attention to the instructions! Go over the annotations of the hits you get, this will save you unnecessary labor. The use of your biological- logic judgment is highly important throughout this exercise.
Cass study:
Bungarus candidus is one of the most dangerous snake species in
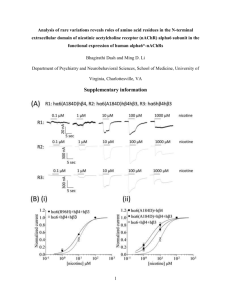
Southeast Asia since no specific antivenom exists to treat envenoming by this species. Death within 30 minuntes after bite suggests presence of a highly lethal postsynaptic neurotoxin in the venom. The toxin is named alpha-bungarotoxin (Mass: 7983.75 Da; LD50: 0.23 microg/g in mice i.p).
Immunoprecipitation was preformed with the alpha-bungarotoxin peptide
[NCBI accession: AAC83982] on protein extracts from human cells. The protein that immunoprecipitated with the alpha-bungarotoxin was sent to mass spectrometry (MS) for identification. The MS analysis yielded the following amino acid sequence:
MEPWPLLLLFSLCSAGLVLGSEHETRLVAKLFKDYSSVVRPVEDHRQV
VEVTVGLQLIQLINVDEVNQIVTTNVRLKQGDMVDLPRPSCVTLGVPL
FSHLQNEQWVDYNLKWNPDDYGGVKKIHIPSEKIWRPDLVLYNNADG
DFAIVKFTKVLLQYTGHITWTPPAIFKSYCEIIVTHFPFDEQNCSMKLG
TWTYDGSVVAINPESDQPDLSNFMESGEWVIKESRGWKHSVTYSCCP
DTPYLDITYHFVMQRLPLYFIVNVIIPCLLFSFLTGLVFYLPTDSGEKMT
LSISVLLSLTVFLLVIVELIPSTSSAVPLIGKYMLFTMVFVIASIIITVIVIN
THHRSPSTHVMPNWVRKVFIDTIPNIMFFSTMKRPSREKQDKKIFTED
IDISDISGKPGPPPMGFHSPLIKHPEVKSAIEGIKYIAETMKSDQESNNA
AAEWKYVAMVMDHILLGVFMLVCIIGTLAVFAGRLIELNQQG
1.
Finding the protein from which this sequence is derived:
1.1.
Go to BLAST and search in Blastp with the a.a sequence above, use refseq and human for elimination. Write the name of the protein and its accession number. What is the length of the full protein?
1.2.
Press the "Links" link on the upper right corner of the refseq entry and then on the "Protein (UniProtKB)". Write the SwissProt accession number.
Tools in Bioinformatics
Class work 7
2.
Inquiring about its function:
2.1.
Find the protein in SwissProt. What is the known function of this protein? What is its known subcellular location?
2.2.
Go to the Gene Ontology section in the SwissProt entry. What is the Molecular function, Cellular component, and related Biological processes of this protein?
2.3.
Identify patterns and motives for the protein: Go to the Prosite server and search for patterns and motives in the protein. Do the results coincide with the information you found in the previous questions? (keep the Prosite window open for later use)
3.
Getting information on the gene from UCSC:
3.1.
Find the genomic location of the gene (chromosome, strand, and coordinates).
3.2.
Play with the zoom to get a good picture of the introns and exons of the gene. How many exons are there?
4.
Finding homologues of the protein:
4.1.
Run blastp with the the protein as query against the refseq database. Answer theoretically: would you use the MS sequence for this search or the refseq/swissprot sequence – explain. Explain whether you should limit your search as in question 1.
4.2.
Provide the accession numbers, description lines, and species name of 10 homologues proteins from different species.
4.3.
For two of your selected hits, provide the NCBI taxon ID of the species.
5.
Multiple sequence alignment:
5.1.
Download the 10 (and the human query) sequences you found in 4.
5.2.
Use MAFFT to align them. Save the alignment file in fasta format.
Tools in Bioinformatics
Class work 7
6.
Conservation:
6.1.
Go to ConSeq and use the MSA (from 5.2.) and choose the human sequence as the query sequence.
6.2.
Are ConSeq's results consistent with Prosite?
6.3.
In the above instructions (questions 5-6), we tried to save time on the expense of the accuracy of the analysis. Write down two ways to improve its accuracy.
Relevant article:
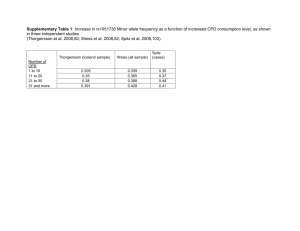
Crystal structure of the extracellular domain of nAChR a1 bound to a-bungarotoxin at 1.94 A° resolution
Cosma D Dellisanti1–3, Yun Yao4, James C Stroud5, Zuo-Zhong Wang4 & Lin
Chen1–3
We determined the crystal structure of the extracellular domain of the mouse nicotinic acetylcholine receptor (nAChR) a1 subunit bound to a-bungarotoxin at 1.94 A° resolution. This structure is the first atomic-resolution view of a nAChR subunit extracellular domain, revealing receptor-specific features such as the main immunogenic region (MIR), the signature Cys-loop and the N-linked carbohydrate chain. The toxin binds to the receptor through extensive protein-protein and proteinsugar interactions. To our surprise, the structure showed a well-ordered water molecule and two hydrophilic residues deep in the core of the a1 subunit. The two hydrophilic core residues are highly conserved in nAChRs, but correspond to hydrophobic residues in the nonchannel homolog acetylcholine-binding proteins. We carried out site-directed mutagenesis and electrophysiology analyses to assess the functional role of the glycosylation and the hydrophilic core residues. Our structural and functional studies show essential features of the nAChR and provide new insights into the gating mechanism.