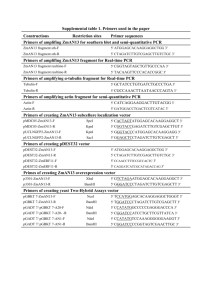
Construction of suicide plasmids required for allelic exchange
advertisement

Supplementary material. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule cluster. Andrea Rahn and Chris Whitfield. The following supplementary material describes: a) the construction of plasmids used for mutation by allelic exchange. b) plasmids used in complementation analyses. c) plasmids containing fragments of the cps promoter region. d) sequences and features of primers used for PCR amplification. 1. Suicide-delivery vector pWQ173. In pWQ173, the sacB gene of pKO3 (Link et al., 1997) was replaced with a different counterselectable marker, pheS (a site-directed mutant of the phenylalanine tRNA synthetase) (Kast and Henneke, 1991; Kast, 1994). Unlike the wildtype PheS, the mutant derivative is promiscuous and allows the incorporation of phenylalanine analogs, such as p-chloro-phenylalanine, into cellular proteins, leading to cell death. To construct pWQ173, the site-directed mutant pheS was PCR-amplified from pKSS (Kast, 1994) using primers SD1 and SD2. The 1 kb fragment was digested with EcoRI and HindIII and ligated to the corresponding sites in pKK223-3 (Amersham Pharmacia Biotech, Inc., Baie d’Urfé, QC). The pheS gene, along with the tac promoter, was removed from this plasmid using BamHI and HindIII and the ends were made blunt using the Klenow fragment of DNA polymerase. This fragment was then ligated to the 3.2 kb HincII fragment from pKO3, effectively replacing sacB. 2. lac::aphA-3 suicide vector, pWQ174. In order to use cps::lacZ fusions, the chromosomal lacZ was first deleted from E. coli isolate B44. The lac operon was PCR- amplified from the chromosome of E. coli strain W3100 using primers lacF and lacR. The 7958 bp PCR fragment was digested with PstI and PinAI and the resulting 5839 bp fragment was cloned into the vector pBAD24 (Guzman et al., 1995), previously cut with PstI and XmaI. An internal 3281 bp fragment containing lacZ was removed from the construct using PvuII and replaced with the kanamycin-resistance cassette (aphA-3) from pYA3265 (Menard et al., 1993). The disrupted lacZ gene was then subcloned into pMAK705 (Hamilton et al., 1989) using PstI and KpnI, creating the suicide plasmid, pWQ174. The construct was used to transform E. coli isolate B44 and allelic exchange was performed as described elsewhere (Hamilton et al., 1989), yielding strain CWG465. E. coli CWG465 was used as the parent strain for many of the other knockout strains described in this work. 3. wbaP::lacZ-aacC1 suicide vectors, pWQ175 and pWQ176. These vectors were constructed using pWQ192, a plasmid derived from the shotgun cloning and sequencing of the K30 cluster. Plasmid pWQ192 includes wbaP, together with ~1.5 kb of flanking sequence. A 1152 bp ScaI/HindIII fragment was subcloned into the HincII/HindIII sites of pMAK705. The lacZ-aacC1 fusion cassette from pAB2002 (Becker et al., 1995) was digested with PstI and ligated into the PstI site within wbaP. This cassette contains a promoterless lacZ, as well as a gentamicin resistance gene, aacC1, which facilitates tracking of the insertion into the chromosome. Two versions of the suicide vector were generated; one with the promoterless lacZ orientated in the direction of cps transcription (pWQ175) and the other in the reverse direction (pWQ176). This allowed for the creation of a wbaP::lacZ fusion strain (CWG466) as well as a strain with the cassette in the reverse orientation on the chromosome (CWG467). 4. wzc::lacZ-aacC1 suicide vector, pWQ177. This construct was based on plasmid pWQ196 (Drummelsmith and Whitfield, unpublished), which contains a 1.7 kb fragment from within the wzc open reading frame. A 375 bp HincII fragment was removed from within wzc and replaced with a SmaI-digested lacZ-aacC1 fusion cassette from pAB2002 (Becker et al., 1995), generating pWQ177. This vector was used to make a wzc::lacZaacC1 fusion strain, CWG468. 5. wzc::aadA suicide vector, pWQ180. This construct was made using the same approach as the vector made for creating the wzc::lacZ fusion vector (pWQ177). Briefly, a 375 bp HincII fragment was removed from pWQ196 and replaced with a polar spectinomycin-resistance cassette (Fellay et al., 1987). 6. rcsB::aadA suicide vector, pWQ178. rcsB was amplified from the B44 chromosome using primers RcsBF and RcsBR, which overlap the open reading frame start and stop codons, respectively. The PCR product was digested with BamHI and HindIII and ligated to the corresponding sites in pMAK705. Following digestion of this plasmid with PstI, the ends were made blunt using T4 DNA polymerase, and then ligated to a SmaI-cut spectinomycin-resistance cassette from pHP45Ω (Fellay et al., 1987). The mutated gene was then removed from the vector as a PvuII fragment and finally ligated into the SmaI site of pWQ173. The final construct was designated pWQ178. 7. galF::lacZ-aacC1 suicide vector, pWQ179. The galF gene was PCR-amplified from the chromosome of strain B44 using the primers AR109 and AR110. The1026 bp fragment was digested with BamHI and cloned into the BamHI site of pWQ173. A 264 bp internal portion of the gene was removed using EcoRV and replaced with the reporter cassette from pAB2002 (Becker et al., 1995). This plasmid, termed pWQ179, was used to make the galF::lacZ fusion strain CWG472. 8. pWQ181 - insertion of a polar cassette downstream of JUMPstart. The B44 cps promoter region was PCR-amplified using primers GalF1 and JD99 yielding a 2.8 kb product. Following digestion with PvuII, the 511 bp fragment carrying the JUMPstart element plus flanking DNA was cleaned and ligated to pWQ173 digested with SmaI. The resultant plasmid was digested with EcoRV, which cuts 82 bp downstream of the JUMPstart element and 145 bp upstream of the first open reading frame of the K30 cps cluster (orfX). The linearized plasmid was ligated to the SmaI-cut spectinomycinresistance cassette (Fellay et al., 1987). 9. rfaH::aadA knock-out vector, pWQ182. The rfaH gene was PCR-amplified from the B44 chromosome using primers AR121 and AR122. The 500 bp product was digested with BamHI and KpnI and ligated to the same sites in pWQ173. This plasmid was then digested with SstII for insertion of the spectinomycin-resistance cassette (Fellay et al., 1987) on a SmaI fragment within rfaH. 10. RcsB overexpression vector, pWQ190. A 662 bp PCR product containing rcsB was amplified using primers RcsBF and RcsBR, digested with BamHI/HindIII and cloned into the same sites in pMALc2x. 11. rfaH expression vector. pWQ191. The rfaH gene was amplified using primers AR126 and AR127 and ligated into the EcoRI and XbaI sites of pBAD24, giving pWQ191. 12. Cloned DNA fragments from the cps promoter region. Smaller fragments in promoter-probe constructs were created by PCR-amplification from a pWQ189 template, using the following primer sets: pWQ242 (primers AR113 and AR135); pWQ243 (AR137 and AR135); pWQ244 (AR137 and AR139); pWQ245 (AR137 and AR141); pWQ246 (AR138 and AR135); pWQ247 (AR140 and AR139). The PCR products were ligated to the SmaI site of pKK232-8 and confirmed using restriction analysis. 13. Sequence analyses. The upstream region of the B44 cps cluster was PCR amplified using primers JD60 and AR110, as well as AR109 and JD98. PCR products of approximately 5.8 kb and 3.9 kb were obtained and sequenced. References: Becker, A., Schmidt, M., Jager, W., and Puhler, A. (1995) New gentamycin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162: 37-39. Fellay, R., Frey, J., and Krisch, H. (1987) Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52: 147-154. Guzman, L.-M., Belin, D., Carson, M.J., and Beckwith, J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121-4130. Hamilton, C.A., Aldea, M., Washburn, B.K., Babtizke, P., and Kushner, S.R. (1989) New method of generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171: 4617-4622. Kast, P. (1994) pKSS - A second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene 138: 109-114. Kast, P., and Henneke, H. (1991) Amino acid substrate specificity of Escherichia coli phenylalanyl-tRNA synthetase altered by distinct mutations. J. Mol. Biol. 222: 99-124. Link, A.J., Phillips, D., and Church, G.M. (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179: 6228-6237. Menard, R., Sansonetti, P.J., and Parsot, C. (1993) Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175: 5899-5906. Primers used for PCR amplification and sequencing. Restriction sites introduced to facilitate cloning are underlined. Lower case letters indicate non-chromosomal sequence. Primer AR103 Sequence AGCGGTTTGAGCTcAGTGGTG Use/Features amplification of stem-loop structure, SstI site; pWQ184 construction. AR104 TTTCCTCtagAATACGCGTCAA amplification of stem-loop structure, XbaI site; pWQ184 construction. AR109 TTGgAtCCAAACAGGTGAAGATG galF amplification, BamHI site; pWQ179 construction. AR110 CCGgAtCCATACGTTTCTTACAATC galF amplification, BamHI site; pWQ179 construction. AR113 GAAATGGAAttCACTGGAGCAC amplification of cps promoter region; forward primer for pWQ242 construction. AR121 GCGCCCgGATcCAACGGATAAG rfaH amplification, BamHI site; pWQ182 construction. AR122 TTGCGGtACcCGGTGTTCTTCA rfaH amplification, KpnI site; pWQ182 construction. AR126 CGGATAgaAtTCATTATGCAATCCTG rfaH amplification, EcoRI site; pWQ191 construction. AR127 AAATGTCtAgACACTGTTTGGGATTG rfaH amplification, XbaI site; pWQ191 construction. AR135 CAGCCGTTGAAGAGAAACCTC amplification of cps promoter region; reverse primer for pWQ242, pWQ243 and pWQ246 construction. AR137 CTCCgAATTcTGACCGAAATT amplification of cps promoter region; forward primer for pWQ243, pWQ244, and pWQ245 construction. AR138 TCAGGaAtTCAGTGCGCTGGT amplification of cps promoter region; forward primer for pWQ246 construction. AR139 ACCAGCGCACTGACTACCTG amplification of cps promoter region; reverse primer for pWQ244 and pWQ247 construction. AR140 TAATGGCCTGACgAATTcTTC amplification of cps promoter region; forward primer for pWQ247 construction. AR141 TATCGACCTGTGGATGAATAATT amplification of cps promoter region; reverse primer for pWQ245 construction. GalF1 GGGCGATCTCTCCGAATACTC amplification of galF-orfX intergenic region. JD60 TTACAACCGCCGTGAAGATG amplification of promoter region. JD62A CTATGAGACGGATGCGCTAATT amplification of wzc internal fragment; pWQ183 construction. JD89 ACGGGCAATAACAGGAAAATA amplification of wzc internal fragment; pWQ183 construction. JD98 GCGGAACCTGACCAAAAGAG amplification of cps promoter region. JD99 GAAAGCAAGCCCAATGTCAC amplification of galF-orfX intergenic region; pWQ181 construction. lacF ACGCCCTGGTTAGTTCAACA amplification of lac operon; pWQ174 construction. lacR ACCCGCGGTCTAATGTTATT amplification of lac operon; pWQ174 construction RcsBF CCggaTcCATGAACAATATGAACGTAATTA rcsB amplification, BamHI TTGC site; construction of pWQ190 and pWQ178. RcsBR AGAaGcTTAGTCTTTATCTGCGGGACTTAA rcsB amplification, HindIII GG site; construction of pWQ190 and pWQ178. SD1 GGAAttCCATGTCACATCTCGC pheS cloning, EcoR1 site; pWQ173 construction. SD2 CATaaGCtTAACCACAGTTCAC pheS cloning, HindIII site; pWQ173 construction.