the DKFZ-HIPO Proposal Form - HIPO
advertisement
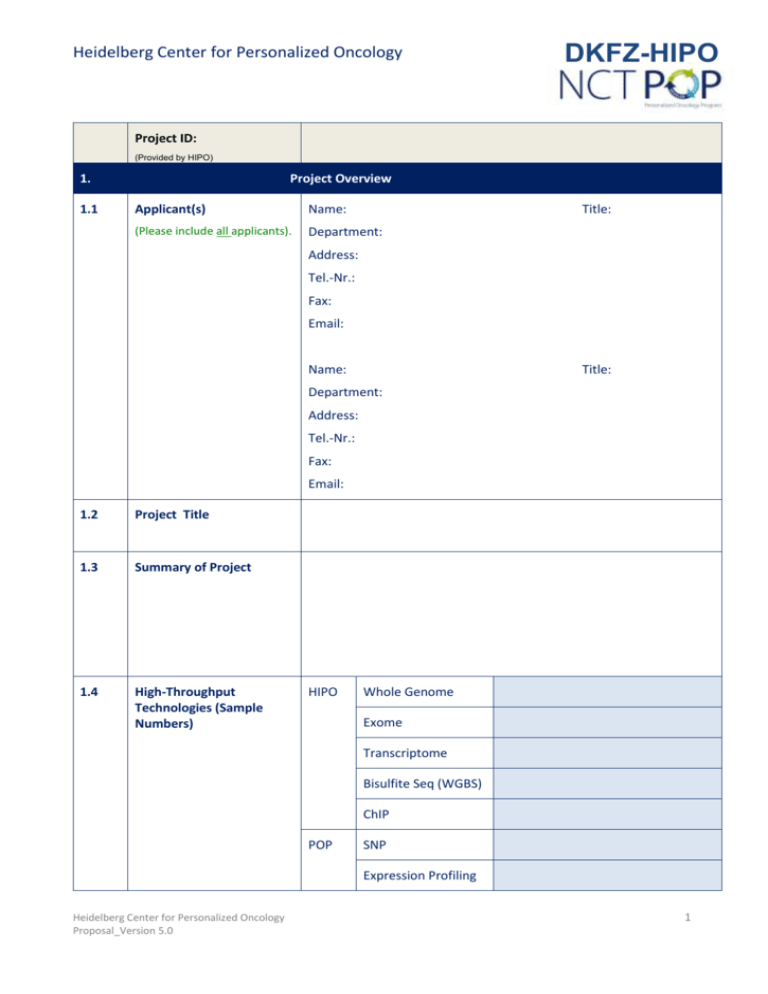
Heidelberg Center for Personalized Oncology DKFZ-HIPO Project ID: (Provided by HIPO) 1. 1.1 Project Overview Applicant(s) Name: Title: (Please include all applicants). Department: Address: Tel.-Nr.: Fax: Email: Name: Title: Department: Address: Tel.-Nr.: Fax: Email: 1.2 Project Title 1.3 Summary of Project 1.4 High-Throughput Technologies (Sample Numbers) HIPO Whole Genome Exome Transcriptome Bisulfite Seq (WGBS) ChIP POP SNP Expression Profiling Heidelberg Center for Personalized Oncology Proposal_Version 5.0 1 Heidelberg Center for Personalized Oncology DKFZ-HIPO 450K Others, please specify: 1.5 Matching Funds from External Sources 2. Project Information 2.1 Background and Significance € (0.5 -1 page) 2.2 Rationale ( 0.5- 1 page) 2.3 Research Design (0.5 - 1 page). Please state your detailed work plan! Heidelberg Center for Personalized Oncology Proposal_Version 5.0 2 DKFZ-HIPO Heidelberg Center for Personalized Oncology 3. Workflow NCT MASTER Informed Consent available for all Samples 3A Sample Preparation 3A.1 Number of Provided Samples or Analytes Yes Others Please specify: Tumor Samples Number of Samples Type (e.g. tissue, DNA, blood, FFPE) Min. Tumor Cell Content Germline Control Number of Samples Type (e.g. tissue, DNA, blood) Others Number of Samples Type (e.g. liquid biopsy) 3A.2 Histological Diagnosis Provided by HIPO/POP Yes No (Institute of Pathology) 3A.3 Analytes Isolation in SPL* required No DNA Number of Samples RNA Number of Samples Others , please specify: Number of Samples *QC and Submission of Samples have to be done exclusively by the Sample Processing Lab (SPL). 3B Molecular and Bioinformatics Analysis 3B.1 Whole Genome Number of Samples Number of Lanes HIPO-STD (3 lanes/sample) other: Person Responsible for Bioinformatic Analysis Heidelberg Center for Personalized Oncology Proposal_Version 5.0 3 DKFZ-HIPO Heidelberg Center for Personalized Oncology 3B.2 Exome Number of Samples Number of Lanes HIPO-STD (3 s.p.l. (tumor), 4 s.p.l. (ctrl)) other: incl. UTR No Kit Yes HIPO-STD (Agilent SureSelect XT Version 5) other: Person Responsible for Bioinformatic Analysis 3B.3 Transcriptome Number of Samples Technique polyA-RNA Seq. other: incl. small RNA Number of Reads No Yes please specify: polyA-STD (40 Mio./ 2,5 samples/lane) other: Person Responsible for Bioinformatic Analysis 3B.4 SNP Genotyping Number of Samples Array-Type Human CytoSNP-12 other: Person Responsible for Bioinformatic Analysis 3B.5 Expression Profiling Number of Samples Array-Type Human HT-12 beadarray other: Person Responsible for Bioinformatic Analysis 3B.6 Epigenomes Number of Samples Technique 450K Whole Genome Bisulfite Sequencing Heidelberg Center for Personalized Oncology Proposal_Version 5.0 4 DKFZ-HIPO Heidelberg Center for Personalized Oncology Number of Lanes (if applicable) Person Responsible for Bioinformatic Analysis 3B.7 ChIP Number of Samples Tumor: Marks H3 (HIPO-Standard Marks are highlighted) H3K4me1 Control: H3K4me3 H3K9ac H3K9me3 H3S10p H3K14ac H3K27ac H3K27me3 H3K36me3 H4 H4K16ac H4K20me3 other (Please specify): TOTAL ChIPs: TOTAL Lanes: Sequencing Type HIPO-STD (50 bp Single Read, 5 s.p.l.) other (Please specify) Person Responsible for Bioinformatic Analysis 3B.8 Others Number of Samples Technique Additional Information (e.g. estimated cost per sample) Person Responsible for Heidelberg Center for Personalized Oncology Proposal_Version 5.0 5 Heidelberg Center for Personalized Oncology DKFZ-HIPO Bioinformatic Analysis 3C Data Access (Bioinformatics, PIs) 3C.1 Request for Data Access Please list all scientists getting access to data (including E-Mail addresses) 3D Clinical Data 3D.1 Patient, Diagnostic, Clinical and Treatment Data Contact Person (including E-Mail address): Follow up: 4. Additional Comments Heidelberg Center for Personalized Oncology Proposal_Version 5.0 6 DKFZ-HIPO Heidelberg Center for Personalized Oncology Signatures □ I have read the HIPO Code of Conduct and agree to the stated terms and conditions. □ I agree to inform the HIPO board of directors about the intention to submit publications 10 days in advance. Publication criteria should be agreed upon within the respective project team. It is expected that the HIPO/POP organizing parties are considered in the usual academic manner. Date: Name: Signature: Date: Name: Signature: □ I am aware that I am responsible to ensure that all patients included in this study have signed the NCT-MASTER consent or an equivalent other consent form that allows for the applied genetic and bioinformatics analyses as well as the storage of data. □ If the study uses a patient consent other than NCT MASTER, I am responsible to ensure the compliance with data storage guidelines, especially time limitations of stored data. I will inform the HIPO Data Management Group (hipo-dtmg@dkfz.de) about approaching deadlines 3 months in advance. Date: Name: Signature: Date: Name: Signature: Heidelberg Center for Personalized Oncology Proposal_Version 5.0 7 Heidelberg Center for Personalized Oncology DKFZ-HIPO Guidelines Please submit 1. Electronically as WORD-document to one of the coordinators (see contact details below) 2. After official acceptance send a signed version of the proposal to one of the coordinators 3. Please note: QC and Submission of Samples have to be done exclusively by the Sample Processing Lab (spl@nct-heidelberg.de) or CHARM (for ChIP) (j.mallm@dkfz-heidelberg.de). Contacts HIPO-Coordinators: POP-Coordinator: Dr. Katja Beck Dr. Janna Kirchhof DKFZ - German Cancer Research Center Abteilung Molekulare Genetik (Prof. Lichter) Im Neuenheimer Feld 280 69120 Heidelberg Dr. Daniela Richter Phone: ++49 6221 42 4579 Fax: ++49 6221 42 4639 Email: j.kirchhof@dkfz.de k.beck@dkfz.de Phone: +49 (0) 6221 56-37819 Fax: +49 (0) 6221 56-6967 Email: daniela.richter@nct-heidelberg.de Heidelberg Center for Personalized Oncology Proposal_Version 5.0 Nationales Centrum für Tumorerkrankungen (NCT) Abteilung Translationale Onkologie (Prof. von Kalle) Im Neuenheimer Feld 460 69120 Heidelberg 8








