Joint UCT/UWC Masters Programme in Structural Biology
advertisement
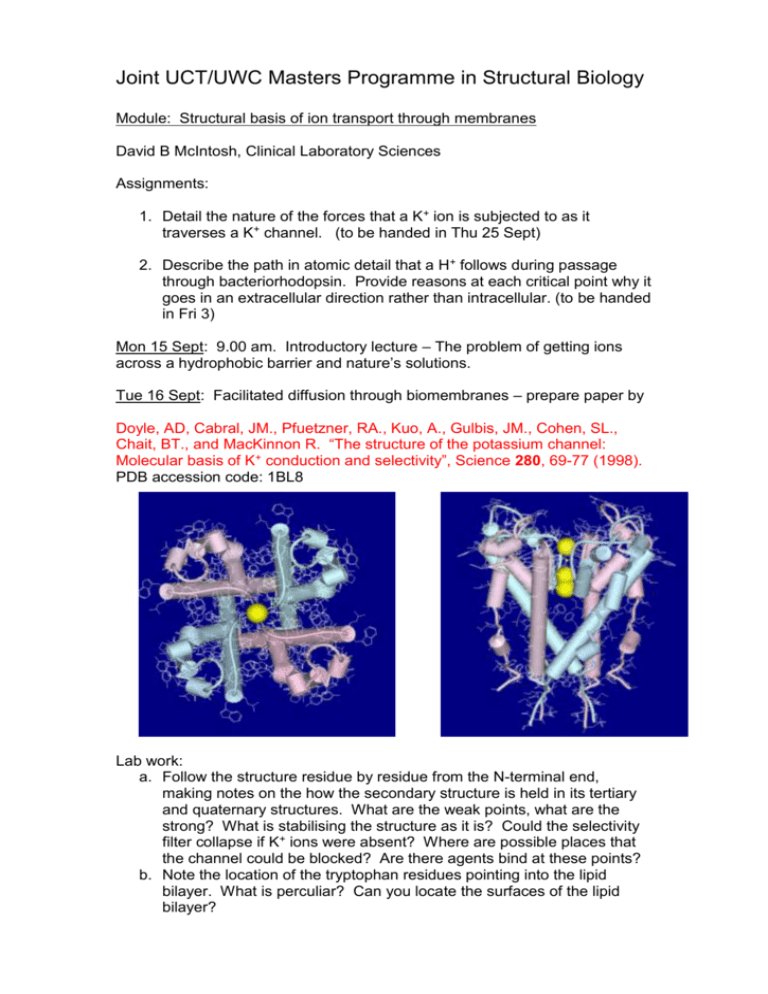
Joint UCT/UWC Masters Programme in Structural Biology Module: Structural basis of ion transport through membranes David B McIntosh, Clinical Laboratory Sciences Assignments: 1. Detail the nature of the forces that a K+ ion is subjected to as it traverses a K+ channel. (to be handed in Thu 25 Sept) 2. Describe the path in atomic detail that a H+ follows during passage through bacteriorhodopsin. Provide reasons at each critical point why it goes in an extracellular direction rather than intracellular. (to be handed in Fri 3) Mon 15 Sept: 9.00 am. Introductory lecture – The problem of getting ions across a hydrophobic barrier and nature’s solutions. Tue 16 Sept: Facilitated diffusion through biomembranes – prepare paper by Doyle, AD, Cabral, JM., Pfuetzner, RA., Kuo, A., Gulbis, JM., Cohen, SL., Chait, BT., and MacKinnon R. “The structure of the potassium channel: Molecular basis of K+ conduction and selectivity”, Science 280, 69-77 (1998). PDB accession code: 1BL8 Lab work: a. Follow the structure residue by residue from the N-terminal end, making notes on the how the secondary structure is held in its tertiary and quaternary structures. What are the weak points, what are the strong? What is stabilising the structure as it is? Could the selectivity filter collapse if K+ ions were absent? Where are possible places that the channel could be blocked? Are there agents bind at these points? b. Note the location of the tryptophan residues pointing into the lipid bilayer. What is perculiar? Can you locate the surfaces of the lipid bilayer? Wed 17 Sept: Details of the selectivity filter – prepare papers by Morais-Cabral et al “Energetic optimisation of ion conduction rate by the K + selectivity filter”, Nature 414, 37-42 (2001). PDB accession codes: 1JVM, 1K4C, and 1K4D Zhou et al “Chemistry of ion coordination and hydration revealed by a K + channel-Fab complex at 2.0 Å resolution” Nature 414, 43-48 (2001). Roux, B., and MacKinnon, R. “The cavity and pore helices in the KcsA K + channel: Electrostatic stabilization of monovalent cations” Science 285, 100102 Labwork: a. View structures and examine the position and coordination of ions b. Locate Asp-80 and Glu-71 c. See if you can see the pinching in of the selectivity filter at Gly-77 in 1K4D Thu 18 Sept: Structural basis of gating – prepare paper by Kuo, A., Gulbis, JM., Antcliff, JF., Rahman, T., Lowe, ED., Zimmer, J., Cuthbertson, J., Ashcroft, FM., Ezaki, T., Doyle, DA. “Crystal structure of the potassium channel KirBac1.1 in the closed state” Science 300, 1922-1926 (2003). PDB accession code: 1P7B Lab work: a. Examine the N-terminal linker and Cterminal domains. Locate the circles of negatively and positively charged residues in the C-terminal domains. What are their roles? Check out the Phe-146. Verify that the inner helix is bent at Gly-134. How are the selectivity filter and pore helices changed from the KscA structure? Significance? b. Write notes on the gating hypothesis. Fri 19 Sept: Introductory lecture: Bacteriorhodopsin - a light powered H+ pump Thu 25 Sept: The ground state structure – prepare paper by Luecke, H., Schobert, B., Richter, H. T., Cartailler, J-P., Lanyi, JK. “Structure of bacteriorhodopsin at 1.55 Å resolution” J. Mol. Biol. 291, 899-911 (1999) PDB accession code: 1C3W Lab work: a. Get familiar with the layout of the 7 helices. See where the phospholipids are located and the position of aromatic residues relative to them. b. Trace the conductance pathway and locate the key residues. Examine in detail the organisation of water molecules under the Schiff base and towards the extracellular surface. Go through the text of the paper with the structure in front of you. Fri 26 Sept: Evolution of activated states – prepare papers by Luecke, H., Schobert, B., Richter, H-T., Cartailler, J-P., and Lanyi, JK. “Structural changes in bacteriorhodopsin during ion transport at 2 Angstrom resolution” Science, 286, 255-260 (1999) PDB accession code: 1CBR and 1C8S (BR and M states of mutant D96N) Lab work: a. In the ground state structure verify that the mutation does not change the organisation of residues and water molecules below the Schiff base. Check the downward orientation of the Schiff base. b. In the M intermediate check the upward orientation of the Schiff base, the single water molecule around Asp-85 and 212. See the downward orientation of Arg-82 and its relationship to Glu-194 and 204. Understand how this allows for the ionisation of the latter. c. Examine the channel above the Schiff base, note the lack of water molecules (just two some distance away). Why is the channel so dehydrated? d. How have the helices changed from the BR state? Concentrate on helices F and G. Lanyi, JK., and Schobert, B. “Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and M2’ intermediates of the photocycle” J. Mol. Biol. 328, 439-450 (2003) PDB accession code: 1O0A (L intermediate) Lab work: a. Colour the BR and L proteins and water molecules differently. Check that the 13 trans to cis isomerisation has taken place, the location of the protonated Schiff base, the water molecules, and the fact that Arg82 has not moved from the BR state. b. Note how helix C has bent inwards. Schobert, B., Brown, LS., and Lanyi, JK. “Crystallographic structures of the M and N intermediates of bacteriorhodopsin: Assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal Schiff base” J. Mol. Biol. 330, 553-570 (2003) PDB accession codes:1P8U (N’ intermediate) Lab work: b Again colour the BR and N’ proteins and water molecules differently. Check the hydration of the region above the Schiff base. Mon 29 Sept: Overview of conformational changes and mechanism of pKa changes - prepare paper by Sampogna and Honig “Environmental effects on the protonation states of active site residues in bacteriorhodopsin” Biophys J. 66, 1341-1352 (1994) Tue 30 Sept: Introductory lecture: Ca2+-ATPase – An ATP driven Ca2+ pump Wed 1 Sept: The E1. Ca2 structure – prepare paper by Toyoshima, C., Nakasako, M., Nomura, H., and Ogawa, H. “Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution” Nature 405, 647-655 (2000) PDB accession code: 1EUL Lab Work: a Become familiar with each domain structure. Locate Asp-351 the residue phosphorylated during the catalytic cycle and note the winding of the highly conserved motif 351DKTGTLT around and behind it. Also find highly conserved motifs 625TGD, 701TGDGVND, and 684K. Speculate on their possible roles. b Find the two Ca2+ and establish how they are ligated. Notice how one ion is coordinated through unravelling of part of a transmembrane helix. The binding and release of Ca2+ from the top (cytoplasm) is ordered. Can you see why? c Locate Phe-487 and Leu-562 in the nucleotide binding domain. The adenine of ATP probably fits between these two residues. Check out Lys-515 which is specifically labelled with FITC. Look for Lys-492 which is specifically photolabelled by TNP-8N3-ATP. Find Arg-678 in the P-domain. Lys-492 and Arg-678 are cross-linked by glutaraldehyde. It is thought to be a zero distance cross-linkage and provides a clue how the N-domain must fit with the P-domain. How close do you reckon the N-domain must approach Asp-351 for phosphorylation to occur. d Locate conserved motif 179TGES in the actuator or A domain. e Try and imagine how phosphorylation of Asp-351 could drive changes at the transport sites. Thu 2 Oct: Structural changes induced by Ca2+ binding – prepare paper by Toyoshima, C., and Nomura, H. “Structural changes in the calcium pump accompanying the dissociation of calcium”. Nature 418, 605-611 (2001) PDB accession code: 1IWO Lab work: a Note the huge change in domain organisation. In particular see where conserved motif 179TGES of the A domain is. Note how much closer the nucleotide site is to Asp-351. b Look what has happened to the membrane helices and Ca2+ sites. Fri 3 Oct: Electrostatic fireworks in the engine room – prepare papers by McIntosh, DB., Woolley, DG., Vilsen, B., and Andersen, JP. “Mutagenesis of segment 487Phe-Ser-Arg-Asp-Arg-Lys492 of sarcoplasmic reticulum Ca2+ATPase produces pumps defective in ATP binding” J. Biol. Chem. 271, 25778-25789 (1996) McIntosh, DB., Woolley, DG., MacLennan, DH., Vilsen, B., and Andersen, JP. “Interaction of nucleotides with Asp351 and the conserved phosphorylation loop of sarcoplasmic reticulum Ca2+-ATPase” J. Biol. Chem. 274, 25227-25236 (1999)





