doc
advertisement

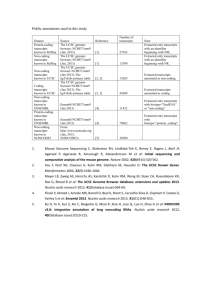
Neema Bhukhan BME 230 Investigating the Importance of Conserved Non-coding Transcripts Abstract: The purpose of this project is to identify the non-coding RNAs using transcription data. I start by first identifying the regions in the ENCODE region of the genome that are actively transcribed by using tiling arrays. Once this information is obtained I subdivide the findings into coding and non-coding transcripts. Then I divide the non-coding transcripts into conserved and not conserved. The evofold track of the UCSC Genome Browser is used to analyze the possible RNA structures of these transcripts excluding any retrotransposons. I searched the ENCODE region and observed patterns of conservation and transcription and analyzed their RNA structure predictions. Introduction: There is little known about what parts of the genome sequence are expressed as mRNA transcripts and whether or not they are functional transcripts. Some areas are conserved but are not transcribed and some sequences are transcribed but not conserved. It is not quite understood what exactly is the relationship between transcription and conservation and if there even is a direct correlation. Does conservation necessarily mean transcription and if so why are there numerous conserved sequences that are non-coding and why are they transcribed? Does non-coding necessarily mean unimportant? The article by Dubchak etal (Dubchak etal 2000) conducted large-scale human-mouse DNA comparisons revealing numerous conserved non-coding sequences, of which only a small percentage are functionally examined. Their inspection revealed almost identical patterns of non-coding sequence conservation in human, dog, and mouse DNA. Of the 14 conserved non-coding sequences found, 2 were determined to be gene regulatory elements. The results they obtained suggest that a large fraction of non-coding elements identified are conserved because of functional constraints. Cawley et al (Cawley et al, 2004) have discovered data that suggest that protein coding and non-coding genes have similar characteristics. There is evidence of the existence of common transcription factors in their promoter regions and the ability to respond to environmental and developmental conditions suggesting they might be controlled by the same transcriptional machinery. These results suggest that non-coding RNAs most likely have important biological functions. The article by Kampa et al (Kampa et al, 2004) points out that the 30,000–40,000 genes in the human genome does not account for any non-coding RNAs. There have been new discoveries of non-coding RNAs such as small nucleolar RNAs, microRNAs, guide RNAs, and antisense RNAs. The addition of these to the gene count would greatly increase the complexity of the human genome. Kapronov et al (Kapronov et al, 2002) have used an empirical approach to create a collection of transcript maps. This approach allows the identification of new regions of transcription, the detection of RNA transcripts with little or no coding capacity, and identification of RNA isoforms of previously annotated genes. Having found new transcripts, they ask why were these transcripts not observed previously and what is their function? They point out that non-coding RNAs are becoming a functional class of transcripts important for splicing, nucleolar and ribosomal structures, telomeric sequence addition, transport and insertion of protein into membranes and down regulation of translation. Characterizing the functions of these transcripts is a task that is making progress and the functions may eventually lead to the discovery of a hidden transcriptome. It is currently difficult to identify the non-coding RNAs, however, as discussed above they are biologically important. Transcription tiling arrays give us information about how often each base in the genome is transcribed under experimental conditions. Using this transcription data I attempt a search for the transcribed non-coding RNA genes by using sequences from the ENCODE region of the human genome. Methods: Using the UCSC Genome Browser I first observed the transfrags track that is based on tiling array data from Affymetrix. The transfrags represent regions of chromosomes 6, 7, 13, 14, 19, 20, 21, 22, X, and Y. Keeping in mind that this track only represents select chromosomes I used the Table Browser to intersect this track with the ENCODE track since I am only interested in the chromosomes of the ENCODE region. I then intersected the ENCODE transfrags with non-coding genes, using the known genes track, to find the non-coding genes that are transcribed. I further divided these non-coding transfrags into those that are conserved and not conserved using the most conserved track. I gathered the statistics of the number of conserved non-coding transcripts and non-conserved non-coding transcripts. Once I had these tracks I excluded the retrotransposons from the transcripts. Then I used the evofold track to analyze the structure predictions of the non-coding transcripts. I picked a couple of the highest scoring, non-coding transcripts to analyze their possible functions. Results: In the ENCODE region I found that 0.87% is conserved non-coding transcripts and 10.48% is non-coding transcripts that are not conserved. When the retrotransposons were excluded the evofold track found 0.09% predictions for the conserved transcripts and 0.65% predictions for the transcripts that are not conserved. Of these evofold predictions I picked the top scoring structures with possibility of functional importance. Figure 1 shows a conserved non-coding structure prediction with a score of 645. Figure 2 shows another conserved non-coding structure prediction with a score of 697. Figure 1: Location of evofold structure prediction with a score 645. Details of structure prediction in attached score645.pdf. Figure 2: Location of evofold structure prediction with a score 699. Details of structure prediction in attached score699.pdf. Discussion: The evofold structure prediction with the score of 645 is located near the FOXP2 gene. The product of the FOXP2 gene is thought to be needed for proper development of speech and language regions of the brain during embryogenesis. The fact that this conserved non-coding transcript is located near the region of this gene suggests the possibility that it may have some functional contribution in the development of this gene. The evofold structure prediction with a score of 699 is located near the end of the same FOXP2 gene suggesting a similar relationship. The results I have obtained suggest conserved non-coding genes are most likely transcribed for a functional reason. Non-coding transcripts should not be disregarded because they can have other relevant functions; the fact that it is not transcribed into a protein does not mean it is unimportant transcript. More recently people have discovered important non-coding transcripts. Perhaps this is why some of these non-coding sequences are conserved. This begins to answer the question of the relationship of conservation and transcription and that there may be some type of correlation. However, there is no direct relationship between the function of the transcripts and if it is coding or non-coding. The results that I have come across go along with the research in this area discussed earlier. Further investigation in this area can lead to the discovery of many new functionally important transcripts that are not currently accounted for in the human genome. References: Dubchak, Inna et al. “Active Conservation of Non-coding Sequences Revealed by Three-Way Species Comparisons.” Genome Research. Vol 10, Issue 9(2000): 1304-1306. Sept, 2000. Kampa, D et al. “Novel RNAs identified from an in-depth analysis of transcriptome of human chromosomes 21 and 22.” Genome Research. Vol 14, Issue 3: 331-342. March, 2004. Cawley, Simon et al. “Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of non-coding RNAs.” Cell. Vol 114, Issue4: 499-509. February, 2004. Kapranov, Philipp et al. “Large-Scale Transcriptional Activity in Chromosomes 21 and 22.” Science.Vol 296, Issue 5569:916-919. May, 2002.