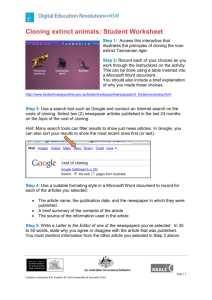
The aim of your research in this class is to clone
advertisement

VDS of the FRI 533566998 Fall 11 *Put your name in the Header of EACH page *Use page numbers Final Research Report This report will take the form of a peer reviewed journal article, just like those that we have been reading in Journal Club. Format: the article should be displayed as TWO COLUMNS. There is an option to do this in WORD. You want your report to look like a journal paper. However, the TITLE and ABSTRACT should not be divided across two columns. See example image of the first page of a journal Page Length: Approximately 3,500 - 5,000 words. Estimate 8-10 pages Single Spaced (11 pt font) with figures and References. Don’t make any pictures bigger than about 1/3 rd of a page TITLE – give a title for your paper, succinct yet conveys what you are doing. Think like an advertising person here. AUTHORS – see a journal paper for example of an author and affiliation list, obviously put yourself there – but also might be a good idea to include the mentors and Dr. B unless you did all the work yourself! Also, if you collaborated with your classmates that worked on the same target – put their names. ABSTRACT – this is a 150-200 word ‘blurb’ about what you did. It is a skeleton of your whole paper and should have the bare essentials necessary to allow a reader to skim it and determine if they care about reading your paper in full or not. We practiced this in class (see Handout for tips) INTRODUCTION (a few paragraphs) Background on your target, motivation for this work (disease) Same info as proposal Be sure to include: Reaction Mechanism image (from BRENDA) for the natural substrate PyMol Image of protein zoomed out (showing overall structure and components – labels would be nice for the intro. Save the zoomed in version of the active site until you talk about virtual screening) Do not include random background pictures from the internet for your disease like you had in the proposal – unless it is a schematic type figure. A regular journal paper would not include these. MATERIALS & METHODS Tell what you did. Will need to explain non-standard methods (e.g. LIC cloning, GOLD) product names (Company Name, City, State) for non-common items Cloning Do not include ‘practice cloning’ on red/green/purple plasmids gene name and size of gene What is the overall purpose of the cloning? What it was cloned into and why? What promoter, tag, antibiotic resistance pNIC-Bsa4 has a his Tag (expression vector) (Company, Location) = (Structural Genomics Consortium, - the one in England) PCR amplification – Primer Overlap (Primary PCR, Secondary PCR, PCR2) primers (list the sequence of the primers) (IDT, Coralville, IA) Final concentration of primers Final MgCl2 concentration that worked Final concentration of dNTPs ng amount of starting plasmid template DNA which polymerase and why? (Company, Location) PCR cycling program (how many seconds and at what temperatures) checked on a 1% agarose gel with Ethidium Bromide pooled samples PCR cleanup kit (Sigma, St. Louis, MO) Cloning method used (Ligation Independent Cloning) 1 VDS of the FRI 533566998 Fall 11 Cohesive end generation with cutting with BsaI first, then T4 DNA Polymerase (What does the T4 Poly do? Which nucleotide did you add?) Annealing and Transformation type of cells (Company, Location) DHFalpha competent cells (NEB, Woods Hollow, MA) Kanamycin and Sucrose plates - why? grow up 5 ml colonies o/n Miniprep to obtain DNA from clones Which kit did you use? (Company, Location) NanoDrop - wavelength used for DNA (Company, Location) verification of cloning Submit to DNA sequencing using which primers (list the sequence of the primers) Expression and purification conditions Transformation of cloned plasmid, pNIC-Bsa4 – gene of interest, containing the T7 inducible promoter for expression of the protein and HIS6 tag, competent cell type - BL21(DE3) - and company, overnight expression final concentrations of antibiotics (Kan or Amp) OD at 600 nm using Red Tide Spec (Company, location) induced with IPTG (spell out) spin down - speed and time which fraction did you save (soluble or pellet) sonicator - type and protocol (pulses, time, etc) spin down again - speed and time, which fraction did you save this time (soluble or pellet) your protein is soluble! overnight freeze? pH adjustment prior to purification Buffer components (Lysis, Wash, Elution) 3 buffer types used Ni-NTA resin for gravity flow column chromatography SDS-PAGE gel to verify purity of elutions and to verify the right size protein is being expressed we make 12% gels (if you used store bought gel it is a 4-20% gel) Centrifugal concentrators (?? MWCO – molecular weight cut off) mention size of your protein (does it get ‘caught’ by the MWCO filter FPLC – briefly mention why it was done, if you used it. quantify protein at 280 nm on a Nanodrop spectrophotometer (compnay, location) storage conditions (% glycerol, temperature) Virtual Screening Briefly - What is the overall purpose of the virtual screening? Say what GOLD is - what it does and how it works - see other papers for brief description of GOLD What does G.O.L.D. stand for, Cite GOLD with (company, location– see the CCDB website) Cite PyMol also Have others done virtual screening on this target – or is your work novel? Protein preparation PDB file name, resolution of Xray crystallography structure from the PDB, how did you set up the protein file to be screened, hydrogens, waters, remove ligand, define Defined active site to ?? Angstroms Don't need to mention Xming - it is just a program that allows you to see a Linux desktop from a Windows desktop Libraries screened, # of ligands, libraries Two step screening process, autoscale Cluster specifics - # of blades, company, operating system, processors, memory, etc GOLD score components – see the VDS stream poster for more info. Aggregate run, Lipinski's rule - define Enzyme assay & Inhibition assay If the substrate is different from the natural one, then show the different Reaction Mechanism (BRENDA). Reaction components: Substrates, buffer conditions, metals. Temperature, time, stop solution, method of measurement (light based or some other method), wavelength, what is the chromophore (i.e. what absorbs the light) 2 VDS of the FRI 533566998 Fall 11 RESULTS Matter-of-factly state and show what your results were. This is where Figures and Tables will go. You do not want to ‘talk’ about results much yet – just state what they were very dryly. The real ‘talking’ about results should go in the Discussion section. Each Figure or Table should have a comprehensive caption explaining what is going on. Reader should not have to reference other parts of the paper in order to understand the Figure. • Cloning gels (PCR, RE check – no plate images of colonies!) • Protein Characterization Gel (attests to purity and quantity) – don’t show Logger Pro image of OD600 protein expression growth • Virtual Results – Aggregate list with Lipinski’s info (denote which ones don’t satisfy the rule) – Note: Lipinski’s original paper should be cited • Small (1.5 inches tall) 2-D images of 3 ligands (the ones you tested in assays – otherwise the top virtual hits that you would have ordered) • 3-D PyMol images zoomed in of active site and docked molecule for 3 different ligands – Show good shot of active site like in the original Research Report write up. • Analyzing your best ligand (PyMol image with explanatory text) – Show your best ligand from screening – binding mode of the ligand into the active site of the protein 1. Potential polar contacts (with hydrophilic amino acids – H bond donors/acceptors) 2. Hydrophobic interactions (with hydrophobic amino acids) • Distances in Angstroms to nearest residues that it may be interacting with (these residues should be shown distinctly when you defined your active site) • Enzyme Assay graphs (vary enzyme, vary susbtrate, vary DMSO) should be in Excel (not Logger Pro) • Inhibition Assay graphs (Absorbance at X nm wavelength [a.u.] on Y-axis and concentration of Inhibitor on x-axis) DISCUSSION (few paragraphs) Talk about your results now and analyze them. Address each figure or table. What were the difficulties in the process? Were the results good/bad? Did the assay work? What were sources of error in your experiments that could make the results difficult to interpret or incorrect? CONCLUSIONS & FUTURE DIRECTIONS (1 paragraph – several sentences) Sum up what has been done (reiterating all that you have just said in a sentence or two) Then propose what your next steps will be (does not need to have specifics but must be detailed enough to show that you know the appropriate things to follow) REFERENCES • Cite at least 5 papers (one should be a review paper and one experimental) ACS (American Chemical Society) bibliography format. Use EndNote Web to do bibliography – Go to FORMAT > BIBLIOGRAPHY > ACS FORMAT > PLAIN TEXT FILE > PREVIEW and then copy and paste to your report at the end. – PubMed, Web of Science (see Spring semester Lit Search Assignment) • You must also cite these electronic resources. Usually there is a webpage telling you how to cite them: – BRENDA http://www.brenda-enzymes.org/information/introduction.php4 – Entrez Gene on NCBI – EXPASY (http://ca.expasy.org/tools/protparam.html) – PDB http://www.rcsb.org/pdb/static.do?p=general_information/about_pdb/policies_references.htm TIPS: Include citations [in brackets] in the body of your paper Don’t use ‘I’ Use third person Use paste tense – almost all of the time Captions should include enough info to let the Figure stand alone *Put your name in the Header of EACH page, Use page numbers Title your filename appropriately – using the standard format that we have been using. UTEID_Name _Date_ EnzymeName_FinalReport.pdf Convert your document to a PDF --------------------------------- Email to Dr. B 3