report 2 guilherme siyang
advertisement

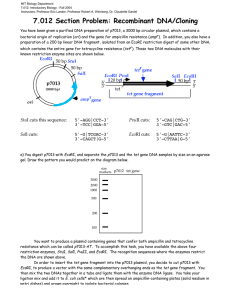
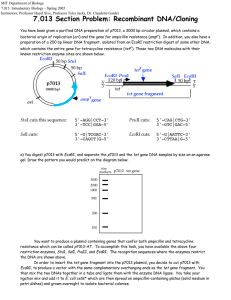
Lab Report 2 by Guiherme and Siyang The objective of the second week is to make us familiar with DNA sciences, which are useful tools for modern biological sciences. There are two main purpose of this lab. The first one is to clone lacZ gene from E. Coli into plasmid pZE21. The second purpose is to measure single cell gene expression using GFP as reporter. Also along the way, we digested -DNA HindIII fragments with EcoRI and use it as DNA ladder. For gene cloning, we first need to extract the lacZ gene (our insert) from a wild type of E. coli (MG1655, GenBank U00096) with PCR. For some unknown reason, the PCR using E. Coli cells does not produce the desired DNA fragment on gel electrophoresis. Instead, we cut out the PCR product using plasmid pZE21-lacZ as template. After purification, we put it into pZE21 vector. pZE21 vector (2948 bp) has moderate copy numbers of 60-70 copies. It has origin of replication colE1, a resistance marker (antibiotic resistance) of Kanamycin, and a regulatory unit (promoter), PLtetO-1, which is repressed by the Tet repressor and induced by tetracycline (ATC). Then we transform the plasmid we made into E. Coli DH5 cells which do not have lac repressor. Also we set up transformations of killer cut to eliminate the effects of relegation and compare without killer cut. “Killer cut” is a restriction digest of the ligation mix using a restriction enzyme that cuts within the starting vector but does not cut the ligated product. This will select against non-recombinants. The rationale for killer cuts is that any contamination with religated starting vector or intact starting vector will be linearized and thus will transform bacteria much less efficiently than the circular intermolecular ligation product. In gene expression measurement, we transform plasmid pZE21-GFP into E. Coli MC4100Z1 cells. MC4100Z1 cells have plasmid for Kanamycin resistance. Then we can measure and compare the induction with ATC. The negative controls are MC4100Z1 which do not have lacZYA but have lacI, tetR and araC, and MC4100Z1pZE21-LacI. The later one is a better control because it has plasmid pZE21-LacI, which is expected to have the same copy number as MC4100Z1-pZE21-GFP, thus better for cancelling auto fluorescence. The positive control is MC4100-pZE21-GFP, which doesn’t have any repression system for GFP expression. From our results, clearly the GFP expression can be induced by ATC. The fold change increases with ATC concentration approximately exponentially. We also performed digestion of lambda DNA and analyzed the fragment size. We start with lambda DNA HindIII digest from Invitrogen. We digested it with EcoRI and run electrophoresis of the DNA before and after digest side by side. From gel electrophoresis, the 4,361bp fragment is almost missing even before EcoRI digestion. This fragment is from the 3’end. It’s likely that we don’t have this fragment in the HindIII digest from Invitrogen. Other fragments corresponds well before and after EcoRI digestion.