a Paradigm for Systems Biology
advertisement
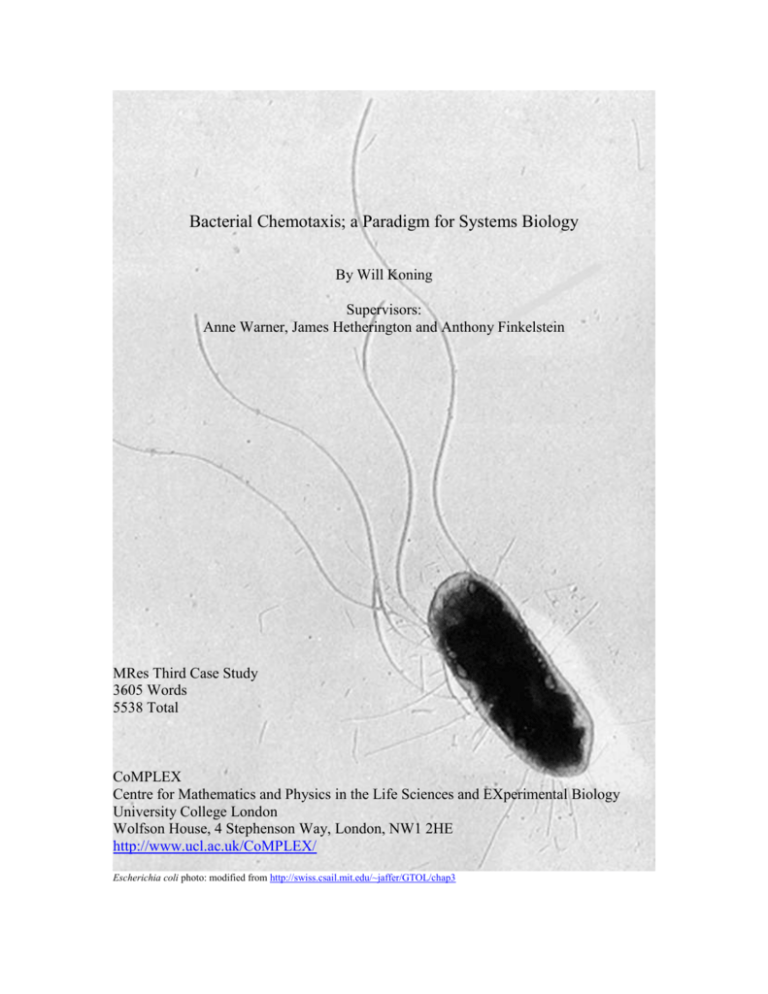
Bacterial Chemotaxis; a Paradigm for Systems Biology By Will Koning Supervisors: Anne Warner, James Hetherington and Anthony Finkelstein MRes Third Case Study 3605 Words 5538 Total CoMPLEX Centre for Mathematics and Physics in the Life Sciences and EXperimental Biology University College London Wolfson House, 4 Stephenson Way, London, NW1 2HE http://www.ucl.ac.uk/CoMPLEX/ Escherichia coli photo: modified from http://swiss.csail.mit.edu/~jaffer/GTOL/chap3 Contents Contents .............................................................................................................................. 2 1 Abstract ........................................................................................................................... 3 2 Introduction ..................................................................................................................... 4 3 Systems Biology ............................................................................................................. 4 4 Bacterial Chemotaxis – Overview .................................................................................. 5 5 Bacterial Chemotaxis – Evolution of Models ................................................................. 8 6 Bacterial Chemotaxis – Paradigm ................................................................................... 8 6.1 Things to consider when modelling biological systems ...................................... 8 6.2 Features of biological systems to consider when developing and testing models ................................................................................................................................... 12 7 Future Work .................................................................................................................. 14 8 Summary ....................................................................................................................... 15 9 Appendix ....................................................................................................................... 16 9.1 Computer models and key experimental studies of bacterial chemotaxis. ........ 16 10 References ................................................................................................................... 20 2 1 Abstract Scientists need to undertake a holistic and integrative approach to understand biological systems. Mass screening technologies are producing large databases of molecular information and methodologies are being developed to integrate this data. Bacterial chemotaxis is a well studied and famous example of systems biology. This essay uses bacterial chemotaxis as a case study to talk about key features of modelling and biological systems. Bacterial chemotaxis has been modelled in silico over the past 12 years and a succession of scientific papers document the development in scope of the models and understanding of the system. 3 2 Introduction “If we hope to understand biology, instead of looking at one little protein at a time, which is not how biology works, we will need to understand the integration of thousands of proteins in a dynamically changing environment” Craig Venter 1999 Craig Venter, while at the helm of Celera Genomics, epitomised the reductionist approach of biology in sequencing the Human Genome but he recognises that scientists need to study the linkages, interactions and processes of these proteins to understand the system. In this essay I give a brief background to systems biology, the study of integrated systems, and then I use bacterial chemotaxis as a case study to explain key features of biological systems and the processes used to model them. 3 Systems Biology Systems biology is the study of integrated biological systems. Most current systems biology projects are grounded on molecular biological knowledge. In 1952 Hodgkin and Huxley modelled action potentials in giant squid (Architeuthis) axons. Their work led Denis Noble to start modelling the electrophysiology of the human heart (Noble, 2002 a and b). Over about 40 years Noble has had tremendous success reproducing the electrophysical dynamics of the heart in silico. Other groups have started modelling other organs; notably the lungs and liver (http://www.bioeng.auckland.ac.nz/projects/lung/pulmonary_models.php; http://pizza.cs.ucl.ac.uk/grid/biobeacon/php/index.php?id=proposal) and the Physiome project has formed to manage many aspects of models of biological systems(http://www.physiome.org) . Biology has progressed rapidly using a reductionist approach but many groups are starting to integrate the vast databases produced using this approach. Biology is moving 4 away from being descriptive and towards theory-based predictive science. Systems biology constructs in silico models that are used to understand, predict and communicate cellular function based on underlying genetic content (Edwards et al. 2001). These models look at different systems using different data and with different scopes but they all integrate relationships of components and have both descriptive and predictive power. Many features of biology can only be understood by looking at the whole system. In this essay I use bacterial chemotaxis as an example of the iterative process of experimentation and modelling in systems biology as it is well understood and shows many key features of the process. 4 Bacterial Chemotaxis – Overview Bacteria can respond to a changing chemical environment by moving. Bacteria sense chemicals and move either towards attractants or away from repellants. Chemotaxis in the bacteria Escherichia coli is perhaps the simplest and best-understood signal transduction network. Chemical receptors are anchored to the cellular membrane. E. coli has five putative chemoreceptor genes (Wadhams and Armitage, 2004). These receptors send chemical messages through the signal transduction network to motors that rotate long flagella that project like hairs from the cell. The flagella motor is the most complex structure in a bacterial cell and in E. coli is the product of the controlled expression of about 50 genes (Wadhams and Armitage, 2004). The flagella are helical and when the motor rotates counter-clockwise the bacteria move forward. When the motor rotates clockwise the flagella loose their helical shape and the bacteria tumble. Bacteria move towards favourable chemical conditions, or away from unfavourable conditions, by engaging in a biased random walk comprising smooth runs and random tumbles. When things are getting better they spend less time tumbling. Bacteria are so small that they cannot detect spatial gradients directly but rather infer them through temporal changes. If conditions are improving (more attractant or less repellent than earlier) the probability of a tumble decreases. 5 Chemical receptors (Trans-membrane receptors; methyl-accepting proteins) and products of genes cheA, cheB, cheR, cheW, cheY, and cheZ (proteins that can exist in a variety of forms) form a network that interacts with the flagellar motor (see Figure 1; Yi et al. 2000; Wadhams and Armitage, 2004). Figure 1. Schematic diagrams of the chemosensory system of Escherichia coli showing interactions (left) and structures (right). (Modified from Wadhams and Armitage 2004, Nature Reviews: Molecular Cell Biology). 6 When the concentration of attractant decreases, the receptor forms a complex with CheW and CheA. CheA then phosphorylates CheY. Then CheY-P (phosphorylated CheY) interacts with a flagellar motor to stimulate tumbling. (Yi et al., 2000; Wadhams and Armitage, 2004). When the concentration of attractant increases, this inhibits the autophosphorylation of CheA, thus reducing the concentration of CheY-P, and lowering the frequency of switching. This allows the motors to spend more time rotating counterclockwise resulting in longer smooth runs (Wadhams and Armitage, 2004). Further complexity arises as CheR-methylates the receptors, which increases CheA activity so more CheY is phosphorylated, thus stimulating tumbling. CheB demethylates receptor balancing CheR and CheZ dephosphorylates CheY-P balancing CheA (see Figure 1; Yi et al. 2000; Wadhams and Armitage, 2004). Adaptation to different concentrations of chemicals occurs because the dynamics of receptor methylation are slow (minutes) relative to CheY phosphorylation (milliseconds) (Yi et al., 2000). To adapt to an attractant the receptors must be more methylated to compensate for saturation of bound attractants. Flagellar motion is normally classified as clockwise or counter-clockwise but the motors not only switch but can also stop and slow down (Wadhams and Armitage, 2004). In short, the chemical receptors assess the concentrations of good and bad chemicals and this information is processed by a collection of molecules that interact to compute whether the bacterium should keep heading in the same direction or whether it should change direction. This computational process works for most different situations. E. coli moves appropriately in response wide range of chemo-attractant concentrations irrespective of ligand concentration because of integral intracellular feedback that insures perfect adaptation (Kitano 2004). The feedback mechanism comprises CheA mediated phosphorylation of CheB, and the dynamics of the stable complex of receptor CheA and CheW, which is methylated by CheR and unmethylated by CheB. Robust adaptation results from CheR working at saturation and CheB only acting on the active form of the methylated complex (independent of the complex’s concentration) (Barkai and Leibler, 1997). One of the reasons this system is so well understood 7 It is that it is easy to locate behavioural mutants, as they spread differently in soft agar, which has assisted the understanding of the system. When these mutants are sequenced, the effect of the mutated gene is demonstrated by the behaviour of the bacteria (natural knock-outs; Alder, 1969; Wadhams and Armitage, 2004). 5 Bacterial Chemotaxis – Evolution of Models Bray et al. produced the first model of bacterial chemotaxis in 1993 for E. coli. Subsequently, a range of models has appeared varying in comprehensiveness and scope (see Appendix). This first model was conceptually simple and comprised elementary chemistry in an homogenous well stirred cell where all the molecules had access to each other. The concentrations of the components and their reaction rates determine the properties of the system (Bray et al., 1993; Weng et al., 1999). The Appendix details the chronology of the models and the remainder of the essay uses the models of bacterial chemotaxis as a case study to talk about systems biology. 6 Bacterial Chemotaxis – Paradigm Bacterial chemotaxis is good example of the iterative process of systems biology at work. Features of bacterial chemotaxis and its models demonstrate key features of systems biology. In this section, I shall discuss things to consider when modelling biological systems and then discuss features of the biological systems themselves that are important to consider. 6.1 Things to consider when modelling biological systems The scope of a model is the particular area of interest. It could be the electrophysiology of a heart or a signal transduction network in a bacterium. It is sensible to ground things at the molecular level as this is where many problems arise, and where vast databases of data are available. The whole system can be built/modelled from these molecular 8 building blocks. Currently there is no reason to explain things in terms of sub-atomic physics. It is important to choose an appropriate level of abstraction based on the aims of the model and the information available. Many people aim to represent the real situation as completely and correctly as possible but there is no need as the purpose is to model rather than simulate (Finkelstein et al. 2004). Trade-offs are made between feasibility, generality, precision and realism. The aim at this stage is to determine what is needed for the model to be useful (Palsson, 2000). Models are simplified versions of organisms. Genomic, biochemical and physiological data is integrated to make in silico models of organisms. These models will have interpretative and predictive capabilities. They may also get things wrong, in which case they are adjusted. All Models are missing features and details and may not have all the links and parameters correct. Bacterial chemotaxis modelling is robust to exact values of kinetic constants(which are hard to determine for an individual cell in vivo) and models are useful with best estimates (Palsson, 2000). A model of E. coli metabolism where rate constants were unknown was useful despite modelling the system based solely on connections between molecules (Edwards et al., 2001). This demonstrates that there are choices in not only what is modelled but also how things are modelled. The type of model used depends primarily on the type of data available. A model is a theoretical image of an object of study so can take many forms. ‘Chemical kinetic models’ model chemical equations using rate constants. They require input of an initial state and they determine amounts of molecules over time in a continuous dynamic fashion. They are not all deterministic (using differential equations; eg. BCT - Bray et al., 1993); some are stochastic (eg. StochSim – Morton-Firth et al., 1999). ‘Discrete circuit models’ use discrete feedback loops with nodes and directed edges. They can be simple (on/off, presence/absence) or include levels. They progress in discrete time steps. All of the bacterial chemotaxis models discussed are chemical kinetic models that tend to increase in complexity chronologically. Barkai and Leibler (1997) used Michaelis-Menten (quasi-steady-state) kinetics for simplicity but found that when cooperative effects in the enzymatic reactions could be included and the robustness remained. BCT uses 60 deterministic rate equations (Lipkow et al., 2005). Solution spaces of a system can be determined which may be quite large depending on the system 9 and the precision of the parameters. Constraints can be used to limit models (diffusion, pressure, systemic stoichiometry, maximum fluxes) by limiting behaviours of many components (Palsson, 2000). Systems biology is a multidisciplinary filed and requires many different skills and backgrounds. The aims of some projects are so ambitious that they require many groups to work together, sharing knowledge expertise and programs. Dennis Bray’s group, which has developed many of the models, highlights this very clearly as it currently comprises eight people, from five different types of department, across four institutions(http://www.anat.cam.ac.uk/~comp-cell/). There is also a not-for-profit organisation, BLAST: Bacterial Locomotion and Signal Transduction, which exists solely for organising meetings in this field (http://www.uic.edu/orgs/blast/), and an alliance, IECA: International E. coli Alliance, which manages E. coli databases (http://www.uni-giessen.de/~gx1052/IECA/ieca.html) demonstrating the collaboration occurring between scientists in systems biology. “If we spoke a different language, we would perceive a somewhat different world” Wittgenstein Scientists speak different languages (e.g. Mathematics and English.) To preserve meaning correctly it is important to standardise the communication, management, experimentation, analysis and modelling of information by developing an ontology; an explicit specification of how to represent objects, concepts and their relationships (Finkelstein et al., 2004). In 1999, a Mars space orbiter was lost because contractor Lockheed Martin Aeronautics used pounds of force in developing a thrusters system while NASA used newtons when they were talking to the orbiter (http://www.cnn.com/TECH/space/9911/10/orbiter.02/). In this case, there should have been an ontology and it should have been followed. In the study of bacterial chemotaxis, most of the models have grown from the first version of BCT (Bray et al., 1993) and, other than through hystoresis, there is no standardisation. In other areas of systems biology software has been developed to 10 standardise processes (e.g. Systems Biology Mark-up Language [SBML]; CellML; and Systems Biology Workbench [SBW]; reviewed in Finkelstein et al., 2004). There are difficulties with collecting data from the literature because of inconsistencies (in researchers, equipment, organisms etc.), and ideally a single strain or cell line should be used under the same conditions to determine all parameters (Kitano 2005). Systems biology involves a parallel and iterative process of experimentation and modelling. The models are used to derive hypotheses about the dynamics of the system that will both test the model and advance the understanding of the system. Failure is an essential part of an iterative process between theory and experiment and often as much is learnt from the failures as from the successes of mathematical models (Noble, 2002b). Bray et al. (1993) noticed the output of their model did not agree with the experimental data for some of the mutants they tested, and these inconsistencies highlighted problems with the model and gaps in the understanding of the system, which they later investigated. Ideally all the components to be modelled are well understood, and their relationships with all other components and the kinetics of these relationships known, but this is unlikely to be the case. High throughput technologies have provided many data about what is in a system but the relationships of these components and their kinetics is normally unknown or based on populations of cells in vitro. Ideally, all data should be from single cells in vivo. Many mathematical modelling methods require more kinetic and enzyme concentration data than is available so estimates are used (Edwards et al. 2001). Data is needed for all the hierarchical levels that the system encompasses. It is important to combine the small components of the system in such a way to try to capture any emergent properties. To keep the models manageable they should be modular and these modules should be integrated (Finkelstein et al. 2004). In the Physiome Project, all the organs will be composed of integrated modules but the organs are themselves modules within the whole body. A model of bacterial chemotaxis will eventually be integrated in to the whole bacterial model that is currently under construction. 11 Ideally, the models will be useful frameworks in other situations (e.g. other cells or other organisms of the same or different species). Many bacterial genomes are sequenced and it appears that the signalling network is common to all but that they differ in its complexity (Wadhams and Armitage, 2004). There is a very similar operon to the one coding for the chemosensory pathway in E. coli, in the bacterium Myxococcus xanthus, where CheA-P phosphorylates either a CheB homologue or a transcription regulator that switches cells from a vegetative state to a sporulation state (Wadhams and Armitage, 2004). This shows genomic data needs to be verified physiologically as homology does not necessitate similar functionality. 6.2 Features of biological systems to consider when developing and testing models Biological systems are robust and stochastic. They are organised spatially and often posess redundancy. These are all important factors to consider in the development of models. It is difficult to determine concentrations and rate constants within an active cell but if many systems are robust and not dependent on exact values, it should be possible to further the understanding of the dynamics of cells without a thorough knowledge of the quantitative data (Barkai and Leibler, 1997). Robust systems can adapt to their environment while being insensitive to specific parameters and degrade noncatastrophically when damaged (Kitano 2002). In bacterial chemotaxis, feedback controls compensate for changes in the initial state of the network (Kitano 2004). In a homogenous environment, steady-state tumbling frequency is independent from ligand concentration, and the rate constants can be changed several fold, resulting in only a few percent difference from perfect adaptation (Barkai and Leibler, 1997). It is possible that some systems are so robust that models give appropriate results despite not only having errors in their rate constants and parameter values but also in their relationships. 12 Models built up of ordinary differential equations are deterministic but molecular activity including gene regulation is essentially stochastic. StochSim includes a stochastic simulation algorithm and is the first model of bacterial chemotaxis to do so (Morton-Firth et al., 1999). Cells are not well mixed homogenous test-tubes but are highly structured and compartmentalised. They are also very crowded. Lipkow et al. (2005) moved away from an electrical circuit idea of interactions and developed Smoldyn to account for cellular architecture, macromolecular crowding, individual molecules that diffuse through the cytoplasm around obstacles (model included 12544 cubic obstacles in lattice taking up 41% of cell volume). Current simulations agree with experiments but go beyond current resolutions possible with existing techniques. Modelling the system with this much detail they were able to show time delays in order of 50ms between motors. It is feasible to simulate all diffusing molecules in the chemotactic pathway (estimated 6000 CheY, 2200 CheYp and 1600 CheZ) and monitor movements and reactions. Chemical receptors are anchored to the membrane (compartmentalisation) and they cluster together (regional organisation) (Weng et al., 1999). Different ends of the receptor are in very different environments where they are exposed to different molecules and conditions. Clustering of the receptors can explain gain in the chemotaxis system (Bray et al., 1998). Molecular scaffolds such as the cytoskeleton provide further regional organisation (Weng et al., 1999). Many biological systems display redundancy (e.g. multiple cells, genes, similar-networks and proteins). Many of the mutants modelled by Bray et al. (1993) still functioned reasonably well despite lacking components of the system. Some components are not essential but simply improve the systems function in some conditions. E. coli may have a streamlined pathway, as other bacteria appear to be more complex and have multiple chemotactic signal transduction pathways or more types of chemosensory protein (Wadhams and Armitage, 2004). These other bacteria may show greater levels of redundancy. 13 7 Future Work Many questions about bacterial chemotaxis in E. coli still remain unanswered – there are a few pages of them on the Computational Cell Biology Group’s website http://www.anat.cam.ac.uk/~comp-cell/Questions.html. More data should be collected from single cells, rather than populations of cells (Mitchison, 2005) and techniques should be developed to do this in vivo. Mass throughput technologies should be developed to calculate kinetic parameters of molecules, similar to the sequencing of genomes. This data should be stored in annotated databases on the web that also define ontologies for particular systems. Most databases store static data but now there is a need to have databases that store dynamic models. Some already do, such as BioModel, a database of annotated published models that was first released on 11 April 2005 (http://www.ebi.ac.uk/biomodels/). Szurmant et al. (2004) found that different species have different proteins in the chemotactic signal transduction network. It would be very interesting to see how they differ and if any function in very different ways. Porter and Armitage (2002) found that CheA proteins in a different species differentially phosphorylate response regulators. Rao et al. (2004) showed it is wrong to make inferences based solely on homology as homologs in both E. coli and Bacillus subtilis tended to function differently and it would be interesting to investigate the evolution of this signalling network in bacteria. Kitano (2004) thinks that mechanisms that incur robustness facilitate evolution, making robustness an inherent property of evolving complex systems and this could be investigated in bacterial species with differing levels of robustness. The understanding of systems biology gained from bacterial chemotaxis can be applied to other systems within E. coli to create a computational model of the whole bacterium. 14 8 Summary There is more to a biological system than the sum of its parts. Emergent properties appear and the system itself is dynamic rather than just its components (Weng et al., 1999). E. coli appears to have a relatively simple system controlling bacterial chemotaxis. Early models of this system were relatively simple, despite involving 60 biochemical reactions, but as the models evolved, they started to include stochastic behaviour of individual molecules existing in a crowded intracellular environment. Notably, the first model was still very useful (Bray et al., 1993). Subsequent models corrected flaws and were more realistic. Adding further detail brings the models closer to the asymptote of an in silico version of the organism itself. However the aim is not to reproduce an exact copy of a system in silico but rather to capture the essence of the system. Biology has had great successes through using a reductionist approach to analyse individual components of systems. It is now going through a revolution as it aims to relate the annotated genome sequence to biological systems (Edwards et al. 2001) and it is moving away from being a descriptive to a predictive science. It is a difficult and timeconsuming process and Denis Noble (2002b) warns researchers not to have unrealistic expectations similar to the ones he faced after the “spectacular success” of the HodgkinHuxley model (action potential in giant squid; 1952). However, with persistence and effort Noble has had even greater success with his heart model. This model is approved for Phase I Drug Trials and Denis Noble predicts that in a few decades there will be models of many organs at this level (http://www.lifescienceit.com/lsitsum04noble.html). 15 9 Appendix 9.1 Computer models and key experimental studies of bacterial chemotaxis. This summary has grown out of two reviews (http://www.anat.cam.ac.uk/~compcell/Review.pdf; Wadhams and Armitage, 2004). The papers chronologically document the development of the understanding of the system. Alder (1969) identified behavioural mutants that spread differently in soft agar due to mutations to their chemotactic genes. Block et al. (1983) investigated clockwise and counter-clockwise spinning over a range of conditions and demonstrated adaptation and robustness of the system. Maddock and Shapiro (1993) identified that chemoreceptors cluster at poles. Bray et al. (1993) made the first comprehensive model of bacterial chemotaxis (BCT program). It is the single most important model as it started the ball rolling and it also demonstrates how much can be achieved in a first sweep. It models the excitation response, but not the adaptation response. The model allows the removal of proteins and when the results were compared with experimental studies, phenotypes of 33 out of 41 mutants accurately reproduced. Bray and Bourret (1995) released an updated version of BCT which incorporates a more realistic model for the formation of the ternary receptor complex. An evolutionary algorithm optimised binding affinities. Vayttaden et al. (2004) ranked models that had been uploaded to 2 public databases and this model came out top equal with six others out of the 244 ranked (no other BCT models were included). 16 Hauri and Ross (1995) constructed a model with kinetic parameters based on experimental measurements that they fine-tuned by an optimisation technique. It was the first model to simulate exact adaptation to both attractant and repellent stimuli. Barkai and Leibler (1997) introduced the concept of robustness in biochemical networks. They showed how it may arise in bacterial chemotaxis through activity-dependent kinetics. Spiro et al. (1997) developed a simplified three-methylation-state model, which they finetuned by trial and error. It simulates ramp, step and saturation responses to an attractant. They used this model to analyse the sensitivity of the system looking at gain, the ratio of stimulus to response. Bray et al. (1998) proposed that sensitivity and gain are because of receptor clustering. The binding of a single ligand to a receptor cluster will affect multiple receptors within that cluster (explaining sensitivity). Subsequent ligand binding to the same cluster will have diminishing effect. As the concentration of ligands increases the proportion of receptors with at least one ligand bound increases rapidly at first but levels off as multiple ligands start binding to the same cluster. This explains the wide range over which the system is sensitive. There is little evidence at present regarding how these lateral interactions occur but it is possibly through changing the charge of all the receptors in the cluster. Levin et al. (1998) further updated the BCT program by incorporating receptormodification reactions that are catalysed by CheR and CheB. They investigated nongenetic individuality through variations in protein expression. Alon et al. (1999) conducted a theoretical and experimental assessment of adaptation and the robustness of the network. 17 Morton-Firth et al. (1999) developed the program StochSim that is the first stochastic model of bacterial chemotaxis. It determines the activity of receptor complexes by calculating the free-energy changes due to both ligand binding and methylation changes. Yi et al. (2000) re-examined the work of Barkai and Leibler (1997) and Alon et al. (1999) with perspective of robust control theory to see how perfect adaptation (appropriate output with high level of robustness) can be achieved with uncertain components in an uncertain environment. Porter and Armitage (2002) demonstrated that different CheA proteins are differentially phosphorylating different response regulators. Shimizu et al. (2003) extended StochSim to model the bacterial membrane in two dimensions so that interactions between neighbouring receptor complexes can be modelled. Mello and Tu (2003) developed a deterministic version of the stochastic model (StochSim) to map the regions of parameter space in which adaptation remains nearperfect. Wadhams et al. (2003) showed that the components of two different chemotactic pathways are separated within the cell. Sourjik and Berg (2004) investigated sensitivity and gain in the chemotaxis pathway and explained it based on the interactions between CheY-P, CheZ and chemoreceptors. Li and Hazelbauer (2004) showed that the relative relationships of signal components maintained over different growth environments. Szurmant et al. (2004) found that different species have different proteins in the chemotactic signal transduction network. 18 Rao et al. (2004) were the first to model chemotaxis in a different bacterial species: Bacillus subtilis. They showed that B. subtilis has two control loops that are not present in E. coli (these feedback loops provide more regulation and robustness). However they do still share certain system design principles. They showed it is wrong to make inferences based solely on homology as homologs in both species tended to function differently. This was done using a combination of experimentation and modelling. Korobkova et al. (2004) investigated intracellular signalling events (as opposed to population events) and found molecular noise (in the amount of CheR produced - which is tuneable) influences behavioural variability. They stated that the design principles of this signal transduction network are found throughout nature and postulated it is likely that molecular noise in other networks is a selectable source of cellular variability. Andrews and Bray (2004) developed the Smoldyn program, which models chemical reaction networks spatially where molecules diffuse freely in three-dimensional space. Reactions can only occur when a pair of reactive molecules collides. This is a general model and is not specific to bacterial chemotaxis. Lipkow et al. (2005) applied Smoldyn to bacterial chemotaxis and modelled the effects of cytoplasmic crowding by excluding spaces. Emonet et al. (2005) developed AgentCell to study the dynamics of stochastic intracellular processes and the behaviour of individual cells. They applied the program to bacterial chemotaxis and implemented the intracellular processes of each bacteria using StochSim (Morton-Firth et al., 1999; Shimizu et al., 2003; Mello and Tu, 2003). They modelled population dynamics of bacteria under stable and attractant gradients where they reproduced the motion of single bacteria and the diffusion of populations. 19 10 References Alon, U., M. G. Surette, N. Barkai and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature. 397:168-171. Andrews, S. J. and D. Bray, 2004. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail Physical Biology 1:137–151. Barkai, N., and S. Leibler. 1997. Robustness in simple biochemical networks. Nature. 387:913-917. Block, S. M., J. E. Segall, and H. C. Berg. 1983. Adaptation kinetics in bacterial chemotaxis. Journal of Bacteriology. 154:312-323. Bray, D., and R. B. Bourret. 1995. Computer analysis of the binding reactions leading to a transmembrane receptor-linked multiprotein complex involved in bacterial chemotaxis. Molecular Biology of the Cell. 6:1367-1380. Bray, D., R. B. Bourret, and M. I. Simon. 1993. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Molecular Biology of the Cell. 4:469-482. Bray, D., M. D. Levin, and C. J. Morton-Firth. 1998. Receptor clustering as a mechanism to control sensitivity. Nature. 393:85-88. Edwards, J. S., R. U. Ibarra RU, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotechnology. 19:111-112. Emonet, T., C. M. Macal, M. J. North, C. E. Wickersham, and P. Cluzel. 2005. AgentCell: a digital single-cell assay for bacterial chemotaxis. Bioinformatics in press. Finkelstein, A., J. Hetherington, L. Li, O. Margoninski, P. Saffrey, R. Seymour, and A. Warner. 2004. Computational challenges of systems biology. Computer. 37:26-33. Hauri, D. C., and J. Ross. 1995. A model of excitation and adaptation in bacterial chemotaxis. Biophysical Journal. 68:708-722. Hodgkin, A. L., and A. F. Huxley 1952. A qulalitative description of membrane current and its application to conduction and excitation in nerve. Journal of Physiology. 117:500-544. Kitano, H. 2002. Systems biology: a brief overview. Science. 295:1662-1664. Kitano, H. 2004. Biological robustness. Nature Reviews: Genetics. 5:826-837. Kitano, H. 2005. International alliances for quantitative modeling in systems biology. Molecular Systems Biology doi: 10.1038/msb4100011. Korobkova, E., T. Emonet, J. M. Vilar, T. S. Shimizu, and P. Cluzel. 2004. From molecular noise to behavioural variability in a single bacterium. Nature 428:574578. Levin, M. D., C. J. Morton-Firth, W. N. Abouhamad, R. B. Bourret, and D. Bray. 1998. Origins of individual swimming behavior in bacteria. Biophysical Journal. 74:175-181. Li, M, and G. L. Hazelbauer. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. Journal of Bacteriology. 186:3687-3694. Lipkow, K., S. S. Andrews, and D. Bray. 2005. Simulated diffusion of phosphorylated CheY through the cytoplasm of Escherichia coli. Journal of Bacteriologyl. 187:45-53. Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 259:1717-1723. 20 Mello, B. A., and Y. Tu. 2003. Perfect and near-perfect adaptation in a model of bacterial chemotaxis. Biophysical Journal. 84:2943-2956. Mitchison, J. M. 2005. Single cell studies and the cell cycle and some models. Theoretical Biology and Medical Modelling. 2:4 doi:10.1186/1742-4682-2-4. Morton-Firth, C. J., T. S. Shimizu, and D. Bray. 1999. A free-energy-based stochastic simulation of the Tar receptor complex. Journal of Molecular Biology. 286:1059-1074. Noble, D. 2002a. Modeling the heart-from genes to cells to the whole organism. Science. 295:1678-1682. Noble, D. 2002b. Modelling the heart: insights, failures and progress. BioEssays. 24:1155-1163. Palsson, B. 2000. The challenges of in silico biology. Nature Biotechnology. 18:11471150. Porter, S. L., and J. P. Armitage. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. Journal of Molecular Biology. 324:35-45. Rao, C. V., J. R. Kirby, and A. P. Arkin. 2004. Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biology. 2:239-252. Shimizu, T. S., S. V. Aksenov, and D. Bray. 2003. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. Journal of Molecular Bilogy. 329:291309. Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proceedings of the National Academy of Sciences 99:7-9. Spiro, P. A., J. S. Parkinson, and H. G. Othmer. 1997. A model of excitation and adaptation in bacterial chemotaxis. Proceedings of the National Academy of Sciences 94:7263-7268. Szurmant, H., T. J. Muff, and G. W. Ordal. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. Journal of Biological Chemistry. 21:21787-21792. Wadhams, G. H., A. V. Warren, A. C. Martin, and J. P. Armitage. 2003. Targeting of two signal transduction pathways to different regions of the bacterial cell. Molecular Microbiology. 50:763-770. Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nature Reviews: Molecular Cell Biology. 5:1024-1037. Wall, M. E., W. S. Hlavacek, and M. A. Savageau. 2004. Design of gene circuits: lessons from bacteria. Nature Reviews Genetics. 5:34-42. Weng, G., U. S. Bhalla, and R. Iyengar. 1999. Complexity in Biological Signaling Systems. Science 284:92-96. Yi, T. M., Y. Huang, M. I. Simon, and J. Doyle. 2000. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proceedings of the National Academy of Sciences 97:5031-5033. 21