Figure 3 (Biorad Laboratories, Inc.)
advertisement
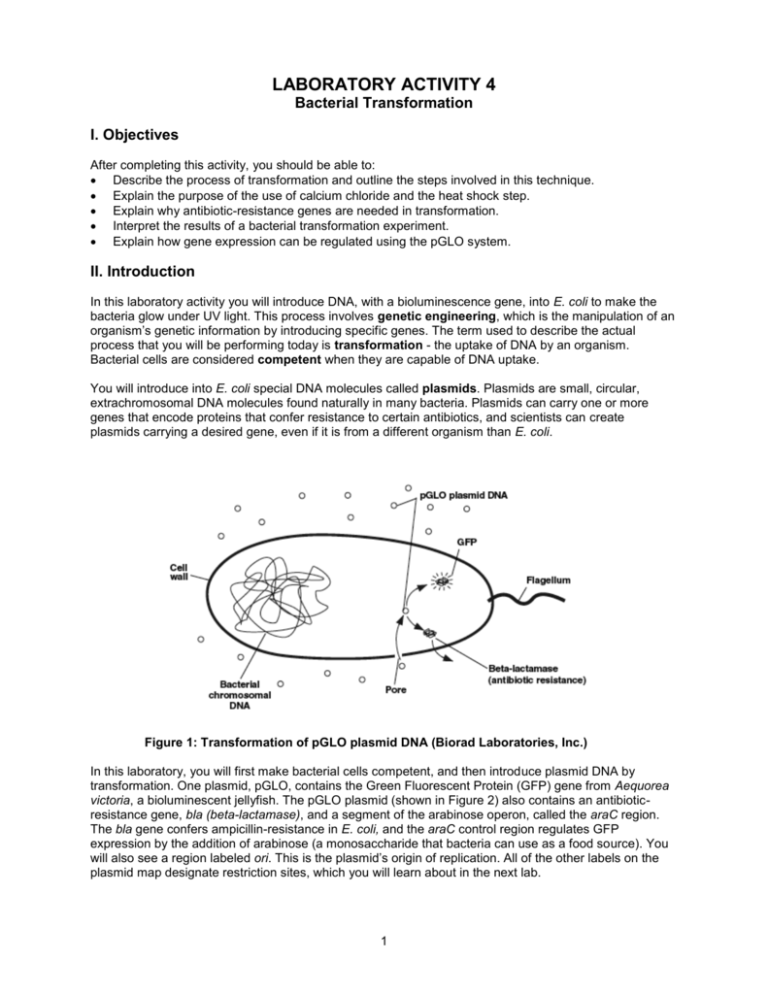
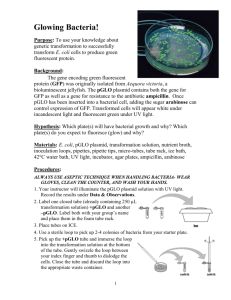
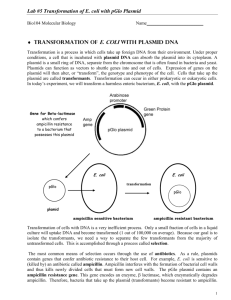
LABORATORY ACTIVITY 4 Bacterial Transformation I. Objectives After completing this activity, you should be able to: Describe the process of transformation and outline the steps involved in this technique. Explain the purpose of the use of calcium chloride and the heat shock step. Explain why antibiotic-resistance genes are needed in transformation. Interpret the results of a bacterial transformation experiment. Explain how gene expression can be regulated using the pGLO system. II. Introduction In this laboratory activity you will introduce DNA, with a bioluminescence gene, into E. coli to make the bacteria glow under UV light. This process involves genetic engineering, which is the manipulation of an organism’s genetic information by introducing specific genes. The term used to describe the actual process that you will be performing today is transformation - the uptake of DNA by an organism. Bacterial cells are considered competent when they are capable of DNA uptake. You will introduce into E. coli special DNA molecules called plasmids. Plasmids are small, circular, extrachromosomal DNA molecules found naturally in many bacteria. Plasmids can carry one or more genes that encode proteins that confer resistance to certain antibiotics, and scientists can create plasmids carrying a desired gene, even if it is from a different organism than E. coli. Figure 1: Transformation of pGLO plasmid DNA (Biorad Laboratories, Inc.) In this laboratory, you will first make bacterial cells competent, and then introduce plasmid DNA by transformation. One plasmid, pGLO, contains the Green Fluorescent Protein (GFP) gene from Aequorea victoria, a bioluminescent jellyfish. The pGLO plasmid (shown in Figure 2) also contains an antibioticresistance gene, bla (beta-lactamase), and a segment of the arabinose operon, called the araC region. The bla gene confers ampicillin-resistance in E. coli, and the araC control region regulates GFP expression by the addition of arabinose (a monosaccharide that bacteria can use as a food source). You will also see a region labeled ori. This is the plasmid’s origin of replication. All of the other labels on the plasmid map designate restriction sites, which you will learn about in the next lab. 1 Figure 2: pGLO plasmid map (Biorad Laboratories, Inc.) As mentioned above, the pGLO plasmid has an AraC region that is important for controlling the expression of the GFP gene. How does it do this? To understand the mechanism, you must first understand how an operon works. Operons are stretches of DNA in bacterial cells that simultaneously control the expression of a group of genes. The genes in the operon generally code for proteins required in the same pathway (e.g., lactose metabolism, tryptophan biosynthesis, arabinose metabolism), and thus need to be turned on at the same time. Operons are composed of several important segments: 1. A promoter - nucleotide sequence where the RNA polymerase binds to begin transcription 2. An operator - nucleotide sequence near or within the promoter, which acts as a “switch,” by controlling access to the promoter. Repressors or activators can bind to the operator to inhibit or stimulate expression of the structural genes, respectively 3. A set of structural genes that are to be transcribed, and later translated The arabinose operon contains 3 structural genes – araA, araB and araD. The protein products of these genes are enzymes that work together to metabolize arabinose into a form that can be used by the cell. Another gene, called AraC, codes for a protein that can bind to the operator and prevent transcription of the structural genes. Arabinose (a monosaccharide) acts as an inducer, or activator. When arabinose is present, it binds to Ara C, causing it to change shape. When this occurs, RNA polymerase can bind to the promoter of the operon, transcription of the enzymes can proceed, and arabinose can be metabolized by the enzymes that are produced. In this case the operon is “on.” If arabinose isn’t present, then the cell doesn’t want to waste the energy to produce enzymes that will break it down. In that case, Ara C prevents the binding of RNA polymerase to the promoter and the operon is off. In the pGLO plasmid, the structural genes of the arabinose operon (araA, B and C) are replaced by the green fluorescent protein (GFP) gene. The Ara C region is still in place, so when arabinose is present, the operon is on, but instead of producing the enzymes to breakdown arabinose, the operon produces the GFP. This process can be visualized because the GFP protein emits a green color under UV light. Figures 3 and 4 illustrate this process. 2 Figure 3 (Biorad Laboratories, Inc.) Figure 4 (Biorad Laboratories, Inc.) 3 III. Procedure Preparing competent E. coli cells 1. Add 20 mL of E. coli HB101 culture to a 50 mL polypropylene centrifuge tube. Spin tube at 3000 x g for 10 minutes in a balanced centrifuge rotor. Cells should form a pellet at the bottom of tube. If cells have not formed a pellet, then repeat centrifuge spin. 2. Carefully pour off the supernatant into a waste beaker containing disinfectant. Invert the tube onto a paper towel to allow the supernatant to drain briefly. 3. With a sterile pipet add 10 mL of ice-cold 50 mM calcium chloride to the tube. Stir the pellet with the end of the pipet and pipet the solution several times to resuspend the cell pellet. The cells should be resuspended completely. 4. Keep the tube on ice for 20 minutes. 5. Spin the cells in a centrifuge for 10 minutes at 3000 x g. 6. Carefully pour off the supernatant into a waste beaker containing disinfectant. Be sure not to disturb the cell pellet. Invert the tube onto a paper towel to allow the supernatant to drain briefly. 7. With a sterile plastic pipet add 2 mL of ice-cold calcium chloride solution to the tube. Resuspend the cells by pipeting the cell solution several times. No cell clumps should be present in the mixture. Keep the cells on ice until you are ready to use them in the next step. Introduction of DNA into E. coli HB101 cells 1. Prepare two microcentrifuge tubes, each containing 200 l of your “competent” HB101 cells. One tube will receive pGLO DNA, and the second tube will be a control, receiving no DNA. Make sue to label these carefully. 2. Add 5 l pGLO plasmid DNA to the cell mixture in the appropriate tube (the one labeled “+pGLO”). Do not add DNA to the other tube (should be labeled “-pGLO” or “no DNA”). Gently tap the bottom of the tube to mix DNA with the cells. Allow cells to sit on ice for 10 minutes. 3. After incubating on ice, heat-shock cells. Bring cells to 370C water bath. Transfer the microcentrifuge tubes from ice immediately to 370C. Allow cells to incubate at 370C for 5 minutes, and immediately transfer tube back to ice. Allow cells to sit on ice for 2 minutes. 4. Add 500 l of LB broth (this is the broth you made in an earlier lab) to each tube of cells and let the cells sit at room temperature for 10 minutes. 5. Obtain 1 LB plate, 2 LB/ampicillin plates and 1 LB/ampicillin + arabinose plate and label them with your name, section number and the date. You will label 1 LB/amp plate and 1 LB/amp/ara plate with “pGLO” and the remaining 2 plates with “no DNA” or “-pGLO.” See the diagram below #7 below to help you correctly label your plates. You always want to label your plates on the bottom, not on the lid, as plates are incubated upside down (since condensation will accumulate on the lids). 6. You are going to make your own bacterial spreader by heating a glass Pasteur pipette with the flame from your Bunsen burner and bending the end until it is at a 90-degree angle. Your instructor will demonstrate this technique. Before plating and spreading your bacterial sample, you will need to sterilize the spreader. To do this, dip the bent end of your spreader into alcohol, and quickly pass the it through your Bunsen burner flame. Let the flame on the spreader burn off the alcohol, and allow the spreader to cool before spreading cells on the agar plate. You can tap the spreader on the side of your agar plate to ensure it is cool enough to use (if it sizzles, it is still too hot). 7. Plate 200 l of each cell mixture onto the appropriate plates (as shown below). Then use the sterile spreaders you made to distribute the liquid evenly on the plate. The cells should be spread across 4 the entire agar surface. Spread each transformation mix on a separate plate, and be sure to sterilize the spreader before using a new transformation mix. 8. Allow the plates to dry, and incubate the plates upside down in a 370C incubator. 9. The plates will be incubated overnight and stored in the refrigerator until next lab session. You will determine your transformation efficiency in the next lab by examining the colonies in a very dark room using a portable long-wave UV light. Post-lab Questions: 1. During this lab, you set up the following 4 plates: I. LB + amp + pGLO DNA II. LB + amp – pGLO DNA III. LB + amp + arabinose + pGLO DNA IV. LB - pGLO On which of the plates (I, II, III, or IV) would you expect to find the following – explain EACH of your answers: a. The most colonies? b. Green colonies? c. No colonies? 2. Which plates should be compared to determine if the transformation was successful? Why? 3. Which plate(s) serve as controls for the transformation? What purpose does the control serve? 4. To genetically transform an entire organism, you must insert the new gene into every cell in the organism. Which organism is better suited for total genetic transformation— one composed of many cells, or one composed of a single cell? 5