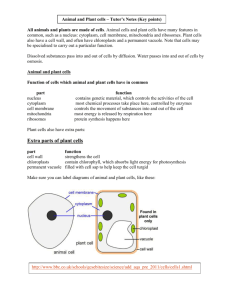
Online supplemental material SUPPLEMENTARY FIGURE

Online supplemental material
SUPPLEMENTARY FIGURE LEGENDS
Supplemental Figure 1: tBid and Bax do not cause liposome fusion. Auto- and cross correlation functions obtained for a 1:1 mixture of green fluorescent liposomes (L
DiO
, ~
40
g/ml lipid concentration) and red fluorescent liposomes (L
DiD
, ~ 40
g/ml lipid concentration) incubated for 1 hour with (A) buffer, (B) 60 nM tBid and 300 nM Bax,
(C) 1 % chloroform or (D) 1.5 % chloroform. Auto-correlation functions obtained from the green and red channels are shown in green and red, respectively, and cross-correlation functions are shown in black. Each curve corresponds to an average of at least 9 experimental auto or cross-correlation curves. The concentration (E,F) and specific brightness (G,H) of the green (E,G) and red (F,H) liposomes in these incubations, as estimated using FIDA, is also shown (mean ± s.e.m., n = 10). These data show that whereas chloroform, as expected, causes liposome fusion (as indicated by a finite crosscorrelation signal, accompanied by a decrease in liposome concentration and an increase in liposome specific brightness), incubation with tBid and Bax does not. Thus liposome fusion does not play any part in the Bax-mediated permeabilization of liposomes lacking tBid shown in Fig. 2, which occurs for lower tBid and Bax concentrations.
Supplemental Figure 2: Binding of tBid to membranes is reversible. (A) Results of gel-filtration experiments for mixture of tBid and liposomes, both fluorescently labeled.
The top panel shows the percentage of liposomes (grey) and tBid (black) found in each fraction when run on the column alone. Lines are Gaussian fits of the data. The middle panel shows the percentage of 200 nM tBid found in each fraction after incubation with
4.75 nM liposomes. The lower panel shows the percentage of tBid in each fraction when the fraction containing the bound protein in the previous elution (fraction 4) is run on the column again. In the last two panels, lines are double Gaussian fit of the data fixing the position of the Gaussians to that found for the data shown in the upper panel. (B)
Schematic outlining the binding equilibria between p7 and tBid, and tBid and the membrane. (C) Fraction of bound tBid or cBid measured by FCS when incubating 20 nM protein with the indicated amount of lipids. The data obtained for tBid was fit assuming a simple equilibrium for the binding of tBid to the membrane ( Eq. 1 , grey line), while the data obtained for cBid was fit assuming two equilibria, one regulating the binding of tBid to the membrane, and one regulating the binding of tBid to the p7 fragment ( Eq. 2 , black line), as shown in panel B (mean ± s.e.m., n=3).
Supplemental Figure 3: Reversible binding of tBid to mitochondria is facilitated by
Mtch2. Mitochondria isolated from mtch2 +/+ and -/- mice
1
were frozen in trehalose as described.
2
For protein to mitochondria FRET, labeling of mitochondria with DiI (Life
Technologies) was carried out as described.
3
Briefly, 2 nM tBid 126C labeled with Alexa
Fluor 488 (tBid
Alexa488
) was incubated with 0.5 mg/ml unlabeled mitochondria (M) with or without Mtch2 for 20 min at room temperature. Mitochondria with bound tBid
Alexa488 were isolated by centrifugation (13000
g for 10 min at 4°C). To observe transfer of tBid
Alexa488
, mitochondria with pre-targeted tBid
Alexa488
were mixed with mitochondria labeled with DiI (M
DiI
) with or without Mtch2 in a ~1:1 ratio for 30 min at 37°C. Bars indicate the percentage of FRET efficiency as a measure of tBid binding to mitochondria.
Final FRET efficiency was calculated as in 3 (mean
s.e.m., n = 3). For each bar, the schematic on the left recapitulates the content of the incubation.
It is clear that tBid
A488
originally bound to +Mtch2 mitochondria successfully migrated to
+Mtch2 M
DiI
(FRET efficiency of 45%), but had notably decreased transfer to –Mtch2
M
DiI
(FRET efficiency of 18%). For –Mtch2 mitochondria, ten fold less tBid
A488
bound to the MOM compared to +Mtch2 mitochondria after incubation (shown previously in
3
).
Therefore, when –Mtch2 mitochondria containing tBid
A488
were incubated with M
DiI
, tBid
A488 had even more robust transfer to +Mtch2 M
DiI
(FRET efficiency of 52%) but only limited transfer to –Mtch2 M
DiI
(FRET efficiency of 25%). Thus, Mtch2 enhances tBid binding if it is in the “accepting” membrane.
Supplemental Figure 4: Binding of Bax to membranes is independent of tBid and cardiolipin. Percentage of FRET efficiency indicative of binding of Bax
DAC
to liposomes
NBD
. 100 nM Bax labeled with the donor, DAC and liposomes containing the acceptor NBD-PE with the indicated lipid composition (See Materials and Methods) were incubated for 30 min at 37°C. FRET efficiency was calculated as explained previously
3
(mean
s.e.m., n = 3).
1. Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M,
Houtkooper RH , et al.
MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nature cell biology 2010, 12 (6) : 553-562.
2. Yamaguchi R, Andreyev A, Murphy AN, Perkins GA, Ellisman MH, Newmeyer
DD. Mitochondria frozen with trehalose retain a number of biological functions and preserve outer membrane integrity. Cell death and differentiation 2007,
14 (3) : 616-624.
3. Shamas-Din A, Bindner S, Zhu W, Zaltsman Y, Campbell C, Gross A , et al.
tBid
Undergoes Multiple Conformational Changes at the Membrane Required for Bax
Activation. The Journal of biological chemistry 2013, 288 (30) : 22111-22127.