Analysis and interpretation of the mutations was conducted
advertisement

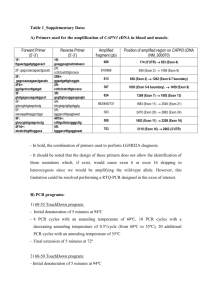
Full clinical description of family 1 Family 1: see Figure 1a. This family lived in France and had both Vietnamese and Martinique origins. The mothers of III.1 and III.12 were of European origin. The proband (III.1) is describedin the main text of the paper. His father (II.1) had complete greying (hair, eyebrows and eyelashes) at the age of 16 years, generalized hypopigmentation of the skin associated with patchy depigmented macules, freckles in sun-exposed regions, lentigines, several cafe-au-lait macules, blue irides with some brown partial heterochromia. Irides were not transilluminable. The fundoscopic examination revealed a slightly diffuse hypopigmentation with small papilla. There was no ocular abnormality ,W Index = 0.85. The audiometric examination was normal. A paternal uncle (II.8) had complete greying (hair, eyebrows and eyelashes) at the age of 10 years, generalized hypopigmentation of the skin associated with freckles and lentigines. He had progressive acquired bilateral perceptive hearing loss of both occupational exposure to intense noises and cochlear sensitivity origins. He complained with photophobia and had blue irides with partial brown heterochromia. The fundoscopic examination revealed sectoral hypopigmentation with retinal pigment clumps. A cousin (III.12) was a 6-year-old boy with onset premature greying, freckles on the face and several cafe-au-lait macules. He complained with photophobia. He had blue irides with partial heterochromia and diffuse retinal hypopigmentation on fundoscopic examination. The audiometric examination showed a small notch on 6 KHz on both ears and tinnitus which could suggest acoustic trauma or some cochlear sensitivity in the context. Another paternal uncle (II.7) had complete greying at the age of 20 years affecting hair, eyebrows and eyelashes, blue irides, diffuse hypopigmentation and freckles. He did not have deafness. He had metastasic thoracic chondrosarcoma. The grandmother (I.2) had Vietnamese origins. She was not clinically examinated but her sons related history of premature greying, blue irides and late deafness. Molecular analysis Sequencing of the MITF-M (melanocytic) isoform coding exons and splice consensus sequences was modified from Tassabehji et al.11 with the use of universal primers UniSeq (N13). PCR was realised in standard conditions (annealing temperature 58° for exons 4, 9A and 9B, 60° for other exons) using primers listed in the Table below. Sequencing was then performed using the UniSeq primers: forward GTAGCGCGACGGCCAGT, reverse CAGGGCGCAGCGATGAC. The absence of total or partial gene deletion was assessed by QMF-PCR (Quantitative Multiplex Fluorescent PCR).12 The coding exons of the MITF-M isoform (including a regulatory region RG that is located in the promoter13-14) and two control fragment of other genes (F9 on chromosome X, DSCR1 on chromosome 21) were amplified in a multiplex reaction using primers listed in the Table below. One primer of each amplicon was labelled with the fluorescent phosphoramidite 6-FAM dye. Amplifications were performed in duplicates in 25 l reactions using the QIAGEN Multiplex PCR kit (Qiagen, France), with 75 ng of genomic DNA, a mix of primers (concentration range 0.15 to 0.6 M), and 5% DMSO. The reaction started with an initial denaturation of 10 min at 95°C followed by 22 cycles at 95°C for 30 sec, 55°C for 30 sec, and 72°C for 45 sec with an increment of 3 sec per cycle, and a final extension of 10 min at 72°C. Then, 4 l of the purified PCR products were processed as previously described. Two normal DNAs (male and female) and one full deletion of MITF were included in each experiment as controls. Primers used in this study PCR-sequence sequence Exon 1M Forward primer Reverse primer GTAGCGCGACGGCCAGTGGATACCTTGTTTATAGTACCTTC CAGGGCGCAGCGATGACAAAAGAGCAGATTTATACTTATTG Exon 2 Exon 3 Exon 4 Exon 5 Exon 6 Exon 7 Exon 8 Exon 9(1) Exon 9(2) GTAGCGCGACGGCCAGTTCTGAAACTCACAAATAACAGCGC CAGGGCGCAGCGATGACTATTCAACAGACAAGTTATTTAGC GTAGCGCGACGGCCAGTCCATCAGCTTTGTGTGAACAGGTC CAGGGCGCAGCGATGACTTTCAGGAAGGTGTGATCCACCAC GTAGCGCGACGGCCAGTAACTAAAGACCATTATTGCTTTGG CAGGGCGCAGCGATGACAGAAAAGAACCCTGGAAACACCTC GTAGCGCGACGGCCAGTATAAATCCTAGAGTAGGATATAGG CAGGGCGCAGCGATGACACTTTGTCTTATCAGGAAATGGAC GTAGCGCGACGGCCAGTTCAAGTCAAATAAGCTTCTGTATG CAGGGCGCAGCGATGACGTAGGAATCAACTCTCCTCTACAG GTAGCGCGACGGCCAGTGTGCTAAATGCATACATGGCACTG CAGGGCGCAGCGATGACTTAGGAATAGAACCAAAGGGAGAG GTAGCGCGACGGCCAGTTTCATTGAGCCTCAAATCCTAAAG CAGGGCGCAGCGATGACCTGTTTCTACTGTCTTGAAGTCGG GTAGCGCGACGGCCAGTAGTCCTCTGTGCTCTGCCTATTTC CAGGGCGCAGCGATGACGCTGCTTGTTTTGGAAGCTC GTAGCGCGACGGCCAGTGGGATCCAAACTGGAAGACA CAGGGCGCAGCGATGACAAGCTAAAGTCTGTGGTGAATTC QMF-PCR RG Exon 1M Exon 2 Exon 3 Exon 4 Exon 5 Exon 6 Exon 7 Exon 8 Exon 9(1) Exon 9(2) F9 DSCR1 Forward primer Reverse primer TTAGATGATGTCTCCTCCAA AAATGTTGATATCAATTTTTCC GGATACCTTGTTTATAGTACCTTC AAAAGAGCAGATTTATACTTATTG TCTGAAACTCACAAATAACAGCGC TATTCAACAGACAAGTTATTTAGC CCATCAGCTTTGTGTGAACAGGTC TTTCAGGAAGGTGTGATCCACCAC AAAGACCATTATTGCTTTGGGTAA AGAAAAGAACCCTGGAAACACCTC ATAAATCCTAGAGTAGGATATAGG TGCACATCACTATATCAATAAC TCAAGTCAAATAAGCTTCTGTATG GTAGGAATCAACTCTCCTCTACAG GGAGAAGTTAATATGCACATGC TTAGGAATAGAACCAAAGGGAGAG TTCATTGAGCCTCAAATCCTAAA TAAGGATTGACTGTGTTAGGATC AGTCCTCTGTGCTCTGCCTATTTC TCCGGGGGACACTGAGGAAAG TCGGTGTCACTGATCCACTC AAGCTAAAGTCTGTGGTGAATTC AAATGATGCTGTTACTGTCTA GAAGTTTCAGATACAGATTTTC GCGACGAGGACGCATTCCAA GTCCTTGTGCGATCACCACA Size 220 bp 270 bp 343 bp 245 bp 259 bp 290 bp 281 bp 300 bp 320 bp 390 bp 369 bp 213 bp 237 bp The underlined sequence corresponds to the universal sequence primers N13. Exon 9(1) stands for the proximal part of exon 9, exon 9(2) the distal part. RG is for regulatory region. Supplementary Figure 3D conformational representation of the basic domain. (a) General representation of Srebp1-A conformation (PDB accession number 1AM9) using the Swiss-Pdb Viewer software. In its bound conformation, the basic domain is an α-helix that is closely inserted within the DNA major groove, residues pointing outside of the α-helix The DNA target is in light blue. One of the two Srebp1-A molecules is in dark blue for clarity, while the other one is showed in yellow and its basic domain in red. In the second picture, a different view of the basic domain α-helix, showing its close insertion within the DNA major groove, residues pointing outside of the α-helix. (b) Sequence alignment between MITF and SREBP1A basic domains. Conserved residues are in red; residues mutated in patients are underligned. (c) These residues are represented in pink in the 3D view. All of them have their side-chain pointing towards DNA. None is located on the externally half the α-helix of the basic domain.