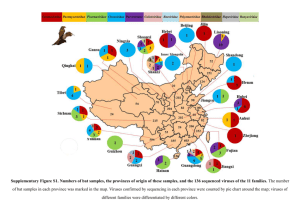
An oasis of viral biodiversity in the Mauritanian desert
advertisement

1 Supplementary Figure 1. Genome coverage for some of the most represented viruses 2 and plasmids. Data were obtained by metagenome read mapping against the specified 3 genomes using GS Reference Mapper with a minimum overlap length of 20 bp and a 4 minimum overlap identity of 95%. 5 6 Supplementary Figure 2. Phylogeny of several ORFs found on the longest contigs from 7 each metagenome. Values on branches represent bootstrap values. A. Phylogenetic 8 reconstruction for ORF 34 on contig 942 of El Berbera, which encodes a hypothetical protein; 9 B. Phylogenetic reconstruction for ORF 7 on contig 439 of Hamdoun, which encodes a 10 possible DNA helicase C. Phylogenetic reconstruction for ORF 41 on contig 353 of 11 Hamdoun, which encodes a bacteriophage protein. 12 13 Supplementary Figure 3. Community structure predicted by PHACCS. Rank-abundance 14 curves representing the community structure and estimates of diversity (R: richness; E: 15 evenness; H’: Shannon-Wiener index of diversity). The abundance of each genotype is plotted 16 versus its abundance rank, with the genotype of rank one being the most abundant. 17 18 Supplementary Figure 4. First two principal coordinates from the principal coordinate 19 analysis of the four samples. The PCA was run by FastUniFrac to visualize the overall 20 patterns of variation between the four samples. 21