Supplementary Materials and Methods (doc 48K)
advertisement

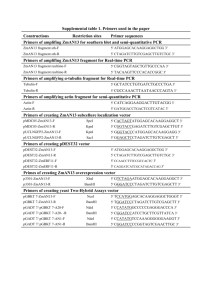
Supplementary Materials and Methods Plasmids and cloning pCDNA3.1myc∆Np63: 2.0 l of cDNA from MCF-7 were used in standard PCR to amplify ∆Np63 for 30 cycles with the following primers: sense 5’CCCAAGCTTAATACGACTCACTATAGGGAGACCATGGAACAAAAACTCAT CTCAGAAGAGGATCTGATGTGTACCTGGAAAACAATG-3’, anti-sense 5’CGGGATCCTCACTCCCCCTCCTCTTTG-3’. HindIII and BamHI restriction sites in sense and antisense primers respectively, are indicated in bold. An Nterminal Myc tag was introduced in each PCR product (sequence highlighted in 5’ primer). ∆Np63 was cloned into pGEM-T vector system I (Promega), the HindIII/BamHI fragments containing the complete ∆Np63 cDNA was then subcloned into the HindIII/BamHI site of pcDNA3 mammalian expression vector (Invitrogen). Identification of the recombinants was also carried out by HindIII/BamHI endonuclease digestion with agarose gel electrophoresis and sequencing. pCMV-Flag∆Np63: pcDNA3.1∆Np63 was used as template for amplification of ∆Np63 by PCR with the following primers sense 5’GGATCCAATACGACTCACTATAGGGAGACCATGGAACAAAAACTCATCT CAGAAGAGGATCTGATGTTGTACCTGGAAAACAATG-3’ and antisense 5’CGGTCGACTCACTCCCCCTCCTCTTTG-3’, BamHI restriction site was introduced in sense primer and Sal I was introduced into antisense primer (both in bold), and myc tag was added at the N terminal of p63 (sequence highlighted in sense primer). PCR products were cloned into pGEM-T vector system I (Promega). The BamHI/ Sal I fragments containing the complete ∆Np63 cDNAs was then subcloned into BamHI/ Sal I sites of pCMV-Flag 2B vector (invitrogen). pGEX6p1-∆Np63: pcDNA3.1-∆Np63 was used as template for amplification of ∆Np63 by PCR with the following primers sense 5’GGATCCATGTTGTACCTGGAAAACAATG-3’ and antisense 5’-CTCGAGTC ACTCCCCCTCCTCTTTG-3’, BamHI restriction site was introduced in sense primer and Xho I was introduced into antisense primer ( both in bold). PCR products were cloned into pGEM-T vector system I (Promega). The BamHI/ Xho I fragments containing the complete ∆Np63 cDNAs was then subcloned into BamHI/ Xho I sites of pGEX 6p1 vector (GE). pcDNA3.1-c-Abl constructs:cDNA from K562 cell line was used for cloning of c-ABL. The following sense primers were used for full length, SH3 and SH3,SH2 c-Abl constructs: Full length c-ABL (1-1149aa) 5’-GGATCC GAGACCATGGAACAAAAACTCATCTCAGAAGAGGATCTGATGGGGCAGC AGCCTGGAAA-3’; SH3 (141-1149aa) 5’-GGATCCGAGACCATGGAAC AAAAACTCATCTCAGAAGAGGATCTGCTGGAGAAACACTCCTGGTA-3’; SH3,SH2 5’-GGATCCGAGACCATGGAACAAAAACTCATCTCAGAAGAG GATCTGGTGGCCGACGGGCTCATCA-3’. Antisense primer for all c-ABL constructs:5’-CTCGAGCTACCTCTGCACTATGTCAC-3’. BamH I and XhoI restriction sites were introduced into sense and antisense primers respectively (both in bold). PCR products were cloned into pGEM-T vector system I (Promega). The BamHI/ Xho I fragments of c-ABL were then subcloned into BamHI/ Xho I sites of pcDNA3.1(+) vector (invitrogen). pEGFP-YAP was described elsewhere (Strano et al., 2001). pCMV-FlagYAP:pEGFP-YAP was used as template for amplification of YAP using 5’GGACGAATTCGATCCCGGGCAGCAG (introducing EcoR1 site) as sense and 5’-TGGCGTCGACC T ATAACCATGTAAGAAAGC (introducing Sal1 site) as antisense primers. PCR product was subcloned in to pCMV-Flag 2B. Mutagenesis Mutants of Np63 and YAP were constructed using QuickChange Sitedirected Mutagenesis Kit (Stratagene) according to manufacturer’s instruction. p63 phosphosite tyrosine (Y) to phenylalanine (F) mutants used pCMVflag-2B-∆Np63 as template for the following primers: ∆Np63 (Y55F) sense 5’-GCGCCCTCGCCCTTTGCACAGCCCAGC-3’ and antisense 5’-GCTGGG CTGTGCAAAGGGCGAGGGCGC-3’; ∆Np63 (Y137F) sense 5'-TCCGCG C CATGCCTGTCTTCAAAAAAGCTGAGCACGTC-3' and antisense 5'-GAC GTGCTCAGCTTTTTTGAAGACAGGCATGGCGCGGA-3’; ∆Np63 (Y308F) sense 5’- CCAGATGATGAACTGTTATTCTTACCAGTGAGGGGCCGTGAG3’ and antisense 5’-CTCACGGCCCCTCACTGGTAAGAATAACAGTTCATC ATCTGG-3’; Triple phosphosite mutant YYYFFF is made from sequential mutation from each p63 phosphosite mutant. pCDNA3.1myc-Np63Y449F(PPXY motif point mutant) used pcDNA3.1 myc ∆Np63 as template for the following primers: sense 5`CACCCCCACCTCCGTTCCCCACAGATTGCAGC–3’ and antisense 5`GCTGCAATCTGTGGGGAACGGAGGTGGGGGTG-3’ Silent p63 phosphosite mutants (Y55F, Y137F and Y308F resistant to siRNA p63) used corresponding p63 phosphosite mutants as template for these primers: sense 5’-TGTTCATCATGCCTCGACTATTTCACGACCCAG GGGC-3’ and antisense 5’-GCCCCTGGGTCGTGAAATAGTCGAGGCA TGATGAACA-3’ pCMV-Flag YAPY357F (YAP-c-Abl phopshosite mutant) was constructed with pCMV-Flag YAP as template with these primers: YAP 357F sense 5’GGACTA AGCATGAGCAGCTTTAGTGTCCCTCGAACCCC-3’ and YAP 357F anti-sense 5’-GGGGTTCGAGGGACACTAAAGCTGCTCATGCTTAGT CC-3’. Letters in primer sequence bolded are sites that were mutated. Mutations were verified by sequencing. Supplemental Figure Legends Figure S1. Mass spectrometric analysis of in vitro (A) and in vivo (B) c-Abl phosphorylation of ∆Np63 reveal multiple tyrosine phosphorylation sites. (A) Recombinant GST ∆Np63 from c-Abl in vitro kinase assays or (B) FLAG∆Np63 immunoprecipitated from c-Abl co-transfected 293 cells were fractionated by SDS-PAGE and ∆Np63 band excised, digested by indicated enzymes and subjected to mass spectrometric analysis as described in Experimental Procedures. Independent peptides containing phosphorylated tyrosine residues are listed. Figure S2. Rescue mutant constructs of ∆Np63 and ∆Np63YYYFFF are resistant to p63 siRNA oligonucleotide. 293 cells were reverse transfected with control or p63 siRNA oligonucleotide and then forward transfected with indicated plasmids. Whole cell lysates were analyzed with indicated antibodies.