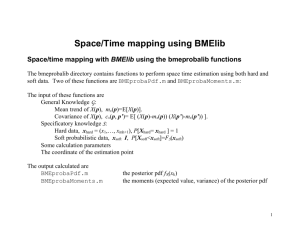
Space/Time mapping with Soft data
advertisement
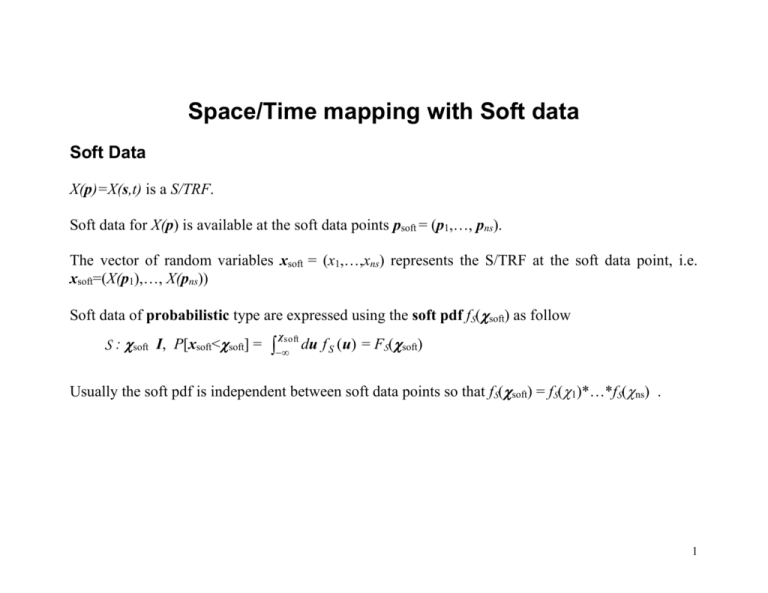
Space/Time mapping with Soft data
Soft Data
X(p)=X(s,t) is a S/TRF.
Soft data for X(p) is available at the soft data points psoft = (p1,…, pns).
The vector of random variables xsoft = (x1,…,xns) represents the S/TRF at the soft data point, i.e.
xsoft=(X(p1),…, X(pns))
Soft data of probabilistic type are expressed using the soft pdf fS(soft) as follow
S : soft I, P[xsoft<soft] =
χsoft
du f S (u) = FS(soft)
Usually the soft pdf is independent between soft data points so that fS(soft) = fS(1)*…*fS(ns) .
1
EXAMPLE:
At point p1, the soft pdf is Gaussian with mean m1=2 and variance 12=3, and at point p2, the soft pdf
is uniform from a2=4 to b2=6. What is the soft pdf for soft = (1 2) ? The answer is as follow
fS(1) =
1 ( m1 ) 2
exp(
)=
2
2 1
12
1
1
( m) 2
exp(
)
6
6
0.5 2 if 4 2 6
fS(2) =
, and
0
otherwise
fS(soft) = fS(1) fS(2)
2
Coding soft data in BMElib
In BMElib, the soft pdf fS(soft) = fS(1)*…*fS(ns) is coded in a discretized form using 4 variables: softpdftype, nl, limi, and
probdens. The reader can type: help probasyntax for a detailed explanation of how these variables work. The first variable
(softpdftype) is an integer taking values 1, 2, 3 and 4. It specifies the type of soft pdf as follows: 1 for histogram, 2 for linear, 3 for
histogram on a regular grid, and 4 for linear on a regular grid. Along each of the ns dimension the univariate pdf fS(i) is defined using
intervals of values for i. The interval limits are specified using the matrix limi, and the value of fS(i) in these intervals is specified
by the matrix probdens.
nl: (nsx1) vector of the number of interval limits. nl(i) is the number of interval limits used to define the soft pdf fS(i) at point
pi
limi: (nsxl) matrix of interval limits, where l is equal to either max(nl) or 3, depending on the softpdftype.
If softpdftype =1 or 2 then limi is a ns by max(nl) matrix, and limi(i,1: nl(i)) are the interval limits for soft data i.
If softpdftype =3 or 4 then limi is a ns by 3 matrix. The interval limits are on a regular grid, and limi(i,1:3) are the lower limit,
increment, and upper limit of the interval limits for soft data i
probdens: (nsxp) matrix of pdf values, where p is equal to either max(nl)-1 or max(nl), depending on the softpdftype.
If softpdftype ==1 or 3 then probdens is a ns by max(nl)-1 matrix. The pdf value is constant in each interval, and probdens (i,
nl(i)-1) are the value of the pdf in each interval.
If softpdftype ==2 or 4 then probdens is a ns by max(nl) matrix. The pdf value varies linearly between interval limits, and
probdens (i, nl(i)) are the value of the pdf at each interval limit.
3
EXAMPLE with softpdftype =1 (soft pdf of histogram type)
>> softpdftype=1;
>> nl=4;
>> limi=[0 2 3 4];
>> probdens=[1 3 2]/7;
>> h=probaplot(softpdftype,nl,limi,probdens);
4
EXAMPLE with softpdftype =2 (soft pdf of linear type)
>> softpdftype=2;
>> nl=4;
>> limi=[0 2 3 4];
>> probdens=[0 4 1 0]/7;
>> h=probaplot(softpdftype,nl,limi,probdens);
5
EXAMPLE with softpdftype =3 (soft pdf of histogram type on regular grid)
>> softpdftype=3;
>> nl=5;
>> limi=[0 1 4];
>> probdens=[1 2 3 2]/8;
>> h=probaplot(softpdftype,nl,limi,probdens);
6
EXAMPLE with softpdftype =4 (soft pdf of linear type on regular grid)
>> softpdftype=4;
>> nl=5;
>> limi=[0 1 4];
>> probdens=[0 3 1 2 0]/6;
>> h=probaplot(softpdftype,nl,limi,probdens);
7
Writing and reading soft data from/to files
The writeProba.m and readProba.m functions allow the user to read and write soft probabilistic
data from/to a file.
Syntax
>>writeProba(cs,isST,softpdftype,nl,limi,probdens,filetitle,datafile);
>>[cs,isST,softpdftype,nl,limi,probdens,filetitle]=readProba(datafile);
EXAMPLE: the following file named ‘somesoftdata.txt’ contains the soft data at two points:
BME Probabilistic data
7
s1
s2
code for the variable (equal to 1)
Type of soft pdf (equal to 1, corresponding to histogram)
number of limit values, nl
limits of intervals (nl values)
probability density (nl-1 values)
1 0.9
1 1 4
0.1 0.3 0.7 1.1
1.0 1.5 0.5
0.1 0.2
1 1 2
0.1 0.3
5.0
Plotting soft data
8
The probaplot.m function, allowing to plot soft data, has the following syntax
>> h=probaplot(softpdftype,nl,limi,probdens,S,idx);
EXAMPLE:
>> [cs,isST,softpdftype,nl,limi,probdens,filetitle]=readProba('somesoftdata.txt');
>> subplot(2,1,1); h=probaplot(softpdftype,nl,limi,probdens,'-',1);
>> subplot(2,1,2); h=probaplot(softpdftype,nl,limi,probdens, '-',2);
9
Generating soft data
The probaGaussian.m and probaUniform.m generate soft data of with Gaussian and uniform distributions,
respectively. For example the following code generate a two soft data points, the first is Gaussian with mean 2 and
variance 3, the second data point is Gaussian with mean 1 and variance 4.
>> [softpdftype,nl,limi,probdens]=probaGaussian([2;10],[3;4]);
>> subplot(2,1,1); h=probaplot(softpdftype,nl,limi,probdens,'-',1);
>> subplot(2,1,2); h=probaplot(softpdftype,nl,limi,probdens, '-',2);
10
% The SRF X(s) is a function of space only in a 2D spatial domain
% This SRF has a mean trend equal to zero, and a covariance
% C(r)=c0*exp(-3r/ar) with c0=1, ar=5
% Additionally we have hard data at two hard data points.
% At s=(.1,4) X(s)=1.2 and at s=(5,2) X(s)=1.7
% And we have soft data (1 , .9) and (2,3).
% We want to estimate the posterior pdf and it's moments at (1,1)
% specify the general knowledge
order=NaN;
% The mean trend is equal to zero
covmodel='exponentialC'; % covariance is exponential, C(r)=c0*exp(-3r/ar)
covparam=[1 5];
% parameters for the covariance model, c0=1, ar=5
% specify the specificatory knowledge
ch=[.1 4;5 2];
% Hard data has two data points, at (.1,4) and (5,2)
zh=[1.2;1.7];
% Value of hard data at (0,4) is 1.2, and at (5,2) it is 1.7
cs=[1 .9;2 3]
% Soft data has two data points, at (1 , .9) and (2,3)
softpdftype=2;
% Soft pdf type=2 correspoinding to linear
nl=[4;3];
% Number of limits for each soft data points
limi=[0 2 3 6;1 2 4 NaN]; % Limits for each soft data points
probdens=[[0 2 10 0]/23;[0 2 0 NaN]/3]; % soft pdf value for each limit value
11
% specify calculation parameters
nhmax=10;
% max number of hard data in estimation neighborhood
nsmax=10;
% max number of soft data in estimation neighborhood
dmax=[100];
% dmax=max spatial search radius for estimation neighborhood
options=BMEoptions;
% Use default options
% specify the coordinate of estimation point
ck=[1 1];
% The estimation point is (1,1)
% calculate BME posterior pdf using BMEprobaPdf
[z,pdf,info]=BMEprobaPdf([],ck,ch,cs,zh,softpdftype,nl,limi,probdens,covmodel,covparam,nhmax,nsmax,dmax,or
der,options);
% calculate moments of BME posterior pdf using BMEprobaMoments
[moments,info]=BMEprobaMoments(ck,ch,cs,zh,softpdftype,nl,limi,probdens,covmodel,covparam,nhmax,nsmax,
dmax,order,options);
expecvalk=moments(:,1)
vark=moments(:,2)
12
13