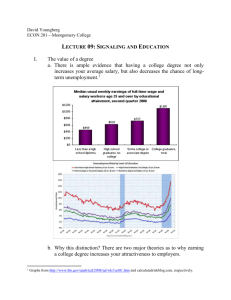
System biological analysis of rebuild signaling pathways
advertisement

5. Stand der Wissenschaft und Technik, bisherige eigene Arbeiten, Patentlage, Wirtschaftliche Bedeutung A. Title of the project System biological analysis of rebuild signaling pathways B. Short Summary Many intracellular signaling elements have been identified in recent years, but their spatial and temporal relationship with each other and with their receptor are poorly understood. Furthermore for any mathematical modeling it is important to obtain also kinetic and quantitative of signaling processes. We have developed a method allowing the reconstitution of mammalian receptors and their signaling cascade in the genetically distant environment of a Drosophila S2 Schneider cell. This synthetic biology approach allows us to freely choose and alter the components of a signaling pathway. Furthermore by controlling the protein expression and function in an inducible fashion we can modify the kinetic and quantitative parameters of signaling pathways. This new approach will be used to verify or falsify mathematical models of signaling. C. Specific Aims Inside the cells signals are processed by a multitude of proteins each of which has its own unique structural and regulatory features as well as different kinetics parameters for signaling. To cope with this complexity, there exist many approaches to generate mathematical models of signaling networks. However, a model is only as good as it can be used to make clear predictions about the outcome of a signaling process inside the cell. To address these aspects of signaling a synthetic-biological approach, namely the rebuilding of signaling pathways can be an important new tool for a better qualitative and quantitative analysis of signaling pathways. By rebuilding a given signaling pathways the experimenter is free to either omit or alter every component of a signaling system. Furthermore by employing inducible techniques for gene expression the experimenter cannot only change the functional but also the quantitative aspect of a signaling network. As most cellular signaling processes require a membrane and a normal cellular environment for their proper function, such a signal reconstitution cannot be done in a test tube. One solution to this problem is the rebuilding of a mammalian signaling pathway in a cell that lacks most 1 components of this pathway. The Drosophila S2 Schneider cells are fulfilling these requirements (Towers and Sattelle, 2002). Indeed, these cells can be efficiently cotransfected in a transient manner with up to 15 plasmids each of which carrying a different gene of a signaling pathway (Rolli et al., 2002). The transient transfection method used in the S2 reconstitution system allows to analyze in a short time frame many mutants of given signaling proteins. Another advantage of the S2 cell system is, that any Drosophila protein that may interfere with the rebuilt signaling pathway, once identified, can efficiently be eliminated by the RNAi technique, which works very efficiently in S2 cells (Caplen et al., 2000). Furthermore each plasmid carries a metallothionein promotor that regulates the expression of the transient transfected gene in an inducible fashion. This allows to study the behavior of a signaling network at different levels of protein production and thus analyse the quantitative aspects of signaling. At present, our experimental read-outs in this system are kinase activation, substrate phosphorylation, protein-protein interaction and signaling complex assembly (Wossning and Reth, 2004). To cope with the complexity of signaling networks, there exist many approaches to generate mathematical models of these signaling. We will apply our S2 Schneider cell Drosophila system to rebuild mammalian signaling pathways, from which mathematical models have been generated and use this system to verify or falsify these theoretical models. The rebuilding approach allows us to omit or change by mutation any component of a given signaling pathway. Furthermore, the inducible fashion, with which we activate gene expression in the S2 cell system allows us not only to change the components, but also the amount of signaling elements in this system. However, right now, in the S2 cells we are only applying regulated promotor for the inducible expression of signaling components. To better study not only the quantitative aspect, but also the kinetic aspect of a signaling pathway it would be important to also have the possibility to induce signaling pathways at a short time frame (in seconds and minutes). To achieve this we will express fusion proteins between the hormone-binding domain of a mutant and Tamoxifen inducible estrogen receptor (ERT2) and kinase domains, which are critical activators of certain signaling pathways. Such ERT2 kinase fusion proteins can be activated in a very short time frame and allow to critically test kinetic parameters of signaling models (Verrou et al., 1999). 2 D. Background and Significance Reverse engineering of intracellular signaling pathways. Crucial cell fate decisions like proliferation, differentiation, survival or death are regulated by a multitude of extra- and intracellular signals. Extracellular signals, such as growth factors are sensed by membrane-bound receptors that translate the extracellular signal into an intracellular activity. Inside the cells signals are processed through multiple pathways resulting in their amplification and regulation by critical feedbacks. The outcome of these signal processes are defined changes in the metabolism, the cytoskeleton and the movement as well as in the transcriptional program of the cell. Due to the concerted efforts of genomic and proteomic approaches many of the components of intracellular signaling pathways have been identified in the last years. However, little knowledge exists about the mechanistic and quantitative aspect of intracellular signaling processes. In Freiburg several research groups have a longstanding interest in studying signaling pathways by either genetic or biochemical means (See study of the insulin receptor pathway, Baumeister group, XX, YY, Signal transduction from the B cell antigen receptor, Reth group). Furthermore, there exists active collaboration with bioinformatic and theoretical groups to generate mathematical models of these signaling pathways (see Timmer group). At present most signaling studies by genetic means are done by the generation of loss of function mutants via the RNAi or knock-out technology. These approaches are very important to identify the functional importance of a given signaling element or a given signaling pathway. However, the mechanistic and the qualitative aspect of signaling pathways as well as the critical feedback loops, which critically determine their outcome, are hard to study by these loss-of-function approaches alone. To address these aspects of signaling a synthetic-biological and reverse engineering approach, namely the rebuilding of signaling pathways from known components can be a powerful research tool for a better and qualitative analysis of signaling pathways (Arkin, 2001; Blake and Isaacs, 2004; Guet et al., 2002; Pawson and Linding, 2005). Indeed the integration of these engineering concepts with computational methods can lead us to a deeper and more abstract understanding of signal transduction systems (Simpson, 2004). 3 Literature: Arkin, A. P. (2001). Synthetic cell biology. Curr Opin Biotechnol 12, 638-644. Blake, W. J., and Isaacs, F. J. (2004). Synthetic biology evolves. Trends Biotechnol 22, 321-324. Caplen, N. J., Fleenor, J., Fire, A., and Morgan, R. A. (2000). dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252, 95-105. Guet, C. C., Elowitz, M. B., Hsing, W., and Leibler, S. (2002). Combinatorial synthesis of genetic networks. Science 296, 1466-1470. Pawson, T., and Linding, R. (2005). Synthetic modular systems--reverse engineering of signal transduction. FEBS Lett 579, 1808-1814. Rolli, V., Gallwitz, M., Wossning, T., Flemming, A., Schamel, W. W., Zurn, C., and Reth, M. (2002). Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop. Mol Cell 10, 1057-1069. Simpson, M. L. (2004). Rewiring the cell: synthetic biology moves towards higher functional complexity. Trends Biotechnol 22, 555-557. Towers, P. R., and Sattelle, D. B. (2002). A Drosophila melanogaster cell line (S2) facilitates post-genome functional analysis of receptors and ion channels. Bioessays 24, 1066-1073. Verrou, C., Zhang, Y., Zurn, C., Schamel, W. W., and Reth, M. (1999). Comparison of the tamoxifen regulated chimeric Cre recombinases MerCreMer and CreMer. Biol Chem 380, 1435-1438. Wossning, T., and Reth, M. (2004). B cell antigen receptor assembly and Syk activation in the S2 cell reconstitution system. Immunol Lett 92, 67-73. 4