predicting the proportion of qtl variation detected from linkage
advertisement

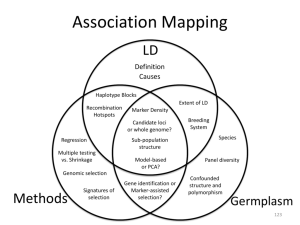
PREDICTING THE PROPORTION OF QTL VARIATION DETECTED FROM LINKAGE DISEQUILIBRIUM BETWEEN GENOTYPED SNP John Bastiaansen Animal Breeding and Genomics Centre, Wageningen University, P.O. Box 338, 6700 AH, Wageningen, The Netherlands For QTL detection and prediction of breeding values used in genomic selection, linkage disequilibrium (LD) is required between genotyped SNPs and QTL. This LD determines the proportion of QTL variation that can be detected by the SNP panel and therefore is an important parameter for the success of genomic selection. Human HapMap data was used to define SNP panels of variable densities and their predictive value for the allelic variation of the non-selected target SNPs was calculated. The predictive value was defined as the average coefficient of multiple determination (R2) of all the target SNPs which served as putative QTL. Multiple genotyped SNPs near the target SNPs explained more of the target SNP variation than the best (highest LD) predicting SNP alone. For a panel with average LD between flanking SNPs of 0.14 (r2) the average maximum LD of target SNPs with a single SNP in the panel was 0.34. With this panel, the average proportion of allelic variation detected by up to 20 genotyped SNPs was 0.62. The relationship between LD in the SNP panel and the proportion of QTL variation detected was found to be similar in populations with greatly different patterns of LD. LD information on the SNP panel alone was found to be a reasonable predictor of detected QTL variation.