Supplementary information (doc 138K)

Materials and methods
Cell culture and reagents
Human breast carcinoma cell lines, MCF-7 and MDA-MB-231 were cultured in
Dulbecco’s modified Eagle’s medium (Gibco, UK) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine at 37°C in a humidified atmosphere of 5% CO2. The non-transformed human breast epithelial MCF-10A cells were cultured in DMEM:F12 (1:1) (Gibco, UK) medium supplemented with 5% horse serum, 10 μg/ml insulin, 20 ng/ml epidermal growth factor (EGF), 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone (all from
Sigma-Aldrich, St Louis, MO, USA), 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. Puromycin (Sigma) was dissolved in DMSO at the concentration of 1 mg/ml as a stock. Kenpaullone (Sigma) was dissolved in DMSO at the concentration of 5 mM as a stock.
Generation of KLF4 knockdown and overexpression stable cell line
Plasmids containing KLF4 specific shRNA (pRetroSuper-KLF4#2, designated as siKLF4) or the control vector (pRetroSuper, designated as siCon) and KLF4 overexpression vector (pBabePuro-KLF4-N, designated as KLF4-N) and the control vector (pBabePuro, designated as Control) were kindly provided by Daniel S. Peeper. pBabePuro-KLF4-N, expressed N-terminal 100 amino acid-fragment of KLF4, can increase KLF4 expression with an unidentified mechanism. 1.6 μg plasmids were
transfected into breast cancer cells using Lipofectamine TM 2000 (Invitrogen, Carlsbad,
CA, USA). 48 h after transfection, cells were trypsinized and reseeded into 10-cm culture plates. Cells were selected with puromycin at a final concentration of 2 μg/ml for 3 weeks.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA was prepared using Trizol Reagent (Invitrogen). First-strand cDNA synthesis was performed using Oligo (dT) and Superscript III reverse transcriptase
(Invitrogen). For reverse transcription of miRNAs, antisense primers of hsa-miR-200c, let-7c and U6 were mixed and cDNA synthesis was performed as described above.
Quantitative real-time PCR was performed as described previously (Ai et al.
, 2004).
Quantitative real-time PCR primer sequences were listed in Supplementary Table 1.
Values were expressed as means ± S.M. of at least three independent experiments.
Western Blot Analysis
Cells with indicated treatment were harvested and lysed in a RIPA buffer. 100 μg protein lysates were separated by 12% SDS–PAGE followed by Western Blot
Analysis as described previously (Ai et al.
, 2004). The antibodies used were as follows: rabbit monoclonal anti-KLF4 (1:1000, Genespin, Milano, Italy), anti-p53
(1:1000, Santa Cruz Biotechnology, CA, USA) and mouse polyclonal anti-α-tubulin
(1:1000, Santa Cruz Biotechnology). Antigen–antibody complexes were detected by
ECL western blotting detection reagent (GE Healthcare, Little Chalfont, UK). Results
shown are representative of three independent experiments.
Isolation of primary breast tumor cells
Primary breast tumor cells from MMTVNeu transgenic mice were digested at 37°C for 2 h in DME-HAM culture medium supplemented with 5% bovine calf serum,
300 U/ml collagenase (Sigma) and 100 U/ml hyaluronidase (Sigma). The resultant suspension was sequentially resuspended in 0.25% trypsin, 5 mg/ml dispase (Roche
Diagnosticsc, Indianapolis, IN, USA) and 0.1 mg/ml DNase (Roche Diagnostics), and
0.64% NH
4
Cl followed by filtration through a 40-μm mesh (Ponti et al.
, 2005). All animals in the experiments were maintained in University of south Carolina School of
Medicine's accredited animal facility. All animal studies were conducted in accordance with NIH animal use guidelines and an approved protocol by the
Institutional Animal Care and Use Committee (IACUC) of University of South
Carolina.
Mammosphere culture
Primary breast tumor cells or MCF-7 cells were suspended in serum-free DMEM:F12
(1:1) culture medium supplemented with 0.4% bovine serum albumin (BSA), 5 μg/ml insulin, 10 ng/ml basic fibroblast growth factor (bFGF), 20 ng/ml EGF (all from
Sigma). Primary breast tumor cells were seeded in 10-cm Ultra-Low Attachment
Surface plates (Corning, NY, USA) at a density of 1 x 10
5
. MCF-7 cells were plated in 96-well Ultra-Low Attachment Surface plates (Corning) at a density of 1000, 500,
200, 100 per well, respectively.
7–10 days later, plates were analyzed for mammosphere formation and were quantified using a Nikon inverted microscope at
10 x magnifications. The data were expressed as means ± S.M. of at least three independent experiments.
Hoechst 33342 staining and sorting
MDA-MB-231 and MCF-7 cells were trypsinized and washed twice in HBSS (Gibco,
UK). 1 × 10
6
cells were resuspended in DMEM growth medium and incubated with
Hoechst 33342 (Sigma) at 5 μg/ml for 90 minutes at 37°C with or without 1 μM
BCRP inhibitor Ko143 (Sigma). After staining, the cells were centrifuged and resuspended in cold HBSS containing 1 μg/ml propidium iodide. SP population analysis and sorting were performed on a MoFlo cytometer (DakoCytomation). The
Hoechst 33342 emission was first split by using a 610-nm dichroic short-pass filter, and the red and the blue emission were collected through 670/30- and 450/65-nm bandpass filters, respectively. Results showed are representative of three independent experiments.
ALDEFLUOR Assay
To assess ALDH activity of the different cell lines, the ALDEFLUOR® assay kit
(StemCell Technologies, Vancouver, BC) was used. MDA-MB-231 and MCF-7 cells were harvested and resuspended in ALDEFLUOR assay buffer at 1 × 10 6
cells/ml. As a negative control, half the sample was transferred to a tube containing 5 μl of the
ALDH inhibitor diethylaminobenzaldehyde (DEAB). 1.5 μl of activated
ALDEFLUOR substrate were added to both samples and incubated at 37°C for 45 minutes to allow substrate conversion. The cells were resuspended in ALDEFLUOR assay buffer and analyzed using the green fluorescence channel (FL1) on a BD
FACSVantage SE (BD Pharmingen, San Diego, CA, USA). Results shown are representative of three independent experiments.
Flow cytometric analysis of CD44+/CD24- and ESA+/PROCR+ population and cell sorting
MCF-7 cells were collected with 0.05% trypsin-EDTA solution and resuspended at 1 x 10 6 cells per ml in 0.5% BSA solution containing diluted anti-human CD24 (FITC) and anti-human CD44 (PE) antibody (BD Biosciences). For identification of
CD44+/CD24- population, samples were analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences). Similar methods were used to analyze the ESA+/PROCR+ population in MDA-MB-231 cells. Sorting of
CD44+/CD24- and CD44-/CD24- cells was performed using a BD FACSVantage SE
(BD Biosciences). The purify of sorted cells was more than 95% by additional flow cytometric analysis. A total of vital 1 x 10
5
cells were collected. Results shown are representative of three independent experiments.
Transfection and dual luciferase assays pGL3-KLF4 promoter plasmid was constructed as described previously (Zheng et al.
,
2009). MCF-7 cells were transfected with 1.6 μg of pGL3-KLF4 promoter plasmid or
IAP promoter (provided by Dr. Chunming Liu) using Superfect transfection Reagents
(from Qiagen). Luciferase activity was measured as described previously (Liu et al.
,
2009).
Scratch assay
MCF-7 or MDA-MB-231 cells were seeded into 6-well plates at a density of 1 × 10
6 cells/well and cultured until confluent in serum-free medium. A pipette tip was used to make a straight scratch. 12 h later, plates were imaged under Nikon microscope using a 10 x Phase contrast objective. Results shown are representative of three independent experiments.
3D Matrigel culture
MCF-7 and MDA-MB-231 cells (1 × 10
4
) were plated onto a 24-well plate coated with a thin layer of growth factor-reduced (GFR) Matrigel Matrix (BD Biosciences).
24 h later, Kenpaullone was added at a final concentration of 1 μM or 5 μM, respectively, for 3 weeks. Phase-contrast photomicrographs were taken by a Nikon inverted microscopy using a 10 x objective. Results shown are representative of three independent experiments.
Adhesion assay
96-well plates were coated with 20 μg/ml fibronectin at 37°C for 1 h and blocked with
0.5% BSA solution for 60 min. 2 × 10 4 cells were added and incubated at 37 o C for 30 min. Cells were fixed with 4% paraformaldehyde and stained with Crystal Violet for
10 minutes. After dissolved in 2% SDS, the absorbance was measured at 550 nm with an ELISA reader (Bio-Tek, Winooski, VT, United States). The data were expressed as means ± S.M. of at least three independent experiments.
Soft agar assay
6-well plates were coated with 1% agar (Cambrex) in growth medium and then overlaid with a suspension of cells (2.5 × 10
3
cells/well) in 0.35% agarose. Plates were incubated for 3 weeks at 37°C and colonies were stained with 0.1% of crystal violet for at least one hour. The number of colonies was determined by counting eight fields of view from triplicate wells for each cell line under a Nikon inverted microscopy using a 10 x objective. The data were expressed as means ± S.M. of at least three independent experiments.
Mouse Tumorigenicity Assay
The siKLF4 and siCon MDA-MB-231 cells were diluted to 5 × 10 5 cells per 20 μl in
PBS. Five NOD/SCID female mice (From the Jackson Laboratory) in the age of 4-6 weeks were injected with the siKLF4 or siCon MDA-MB-231 cells subcutaneously into the mammary fat pad number four according to standard injection procedures.
Once tumors are palpable (about two weeks after injection of tumor cells), primary tumor growth rates were analyzed by measuring tumor length ( L ) and width ( W ), and
tumor volume are calculated using the formula: ( L × W 2 )/2. Mice were sacrificed in a humane manner when the tumor size exceeds 25 mm in diameter in either direction.
Statistical analysis
Two-way ANOVA and two-tailed t -tests were employed to analyze the in vitro data using Prism 5.0 software (GraphPad). P < 0.05 was defined as statistically significant.
Supplementary references
Ai W, Liu Y, Langlois M, Wang TC (2004). Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J Biol Chem 279: 8684-93.
Liu G, Zheng H, Ai W (2009). C-terminal binding proteins (CtBPs) attenuate
KLF4-mediated transcriptional activation. FEBS Lett 583: 3127-32.
Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D et al (2005).
Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 65: 5506-11.
Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, Liu C et al (2009). KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol
296: G490-8.
Supplementary Table
Primers for quantitative real-time PCR and semi-quantitative RT-PCR
Sense Antisense
GAPDH
KLF4
Notch1
5’-TTCGACAGTCAGCCGCATCTTCTT-3’ 5’-CAGGCGCCCAATACGACCAAATC-3
5’-GAAATTCGCCCGCTCCGATGA-3’ 5’-CTGTGTGTTTGCGGTAGTGCC-3’
5’-GCAGACTATGCCTGCAGCTG-3’
5’-ATTGCAGTGTCGAGATGGCT-3’
5’-ACGGGTCAGCCTGCTGGCAC-3’
5’-CTGAGGCACATCGGCACGTG-3’
Notch2
Jagged1
5’-TCGCTGTATCTGTCCACCTG-3’ hsa-miR-200c
5’-CTCGTCTTACCCAGCAGTGT-3’ hsa-let-7c
5’-TTGAGGTAGTAGGTTGTATGGTT-3’
5’-AGTCACTGGCACGGTTGTAG-3’
5’-GTCATCATTACCAGGCAGTATTAG-3’
5’-GGAAAGCTAGAAGGTTGTACAG-3’
U6
Oct4
Nanog
Actin
5’-CTCGCTTCGGCAGCACA-3’
5’-GTGGAGGAAGCCGACAACAATG-3’
5’-AGAAATCCCTTCCCTCGCCA-3’
5’-GAAAATCTGGCACCACACCT-3’
5’-AACGCTTCACGAATTTGCGT-3’
5’-GCCTCATACTCTTCTCGTTGG-3’
5’-TGGTAGAAGAATCAGGGCTG-3’
5’-GGCCGGACTCGTCATACTC-3’
Supplementary Figure legends
Supplementary Figure 1. KLF4 expression was examined in KLF4 knockdown and overexpressing stable cell lines. (a). KLF4 expression in control (designated as siCon) and KLF4 knockdown (designated as siKLF4) cells were detected by quantitative real-time PCR (left) and Western blot (right). * P < 0.05 vs siCon group. Tubulin was used as an internal control. (b). Similar to (a) except control (designated as Control) and KLF4-overexpressing cells (designated as KLF4-N) were used.
Supplementary Figure 2. KLF4 expression in CD44+/CD24- and CD44-/CD24- populations were examined in control (siCon) and KLF4 knockdown (siKLF4) cells respectively. 1 x 10
5
cells in each population were sorted and quantitative real-time
PCR (a) and Western blot (b) were performed to detect KLF4 expression. * P < 0.05 vs CD44+/CD24- group.
Supplementary Figure 3. Overexpression of KLF4 increased the population of stem/progenitor cells in MCF-7 and MDA-MB-231 background. (a). Flow cytometric analysis of CD44+/CD24- population in KLF4 overexpression cells in MCF-7 strain background. (b). SP in MCF-7 stable cells (KLF4-N and Control) was determined by
Hoechst 33342 Efflux Assays. 1 x 10 6 MCF-7 stable cells were incubated with 5 µM
Hoechst 33342 in the absence (top) or presence (bottom) of 1 μM BCRP inhibitor
Ko143. SP cells were analyzed by flow cytometry as described in Materials and
Methods. ALDH activity in MCF-7 (c) and MDA-MB-231 (e) stable cells was
assessed by utilizing the Aldefluor assay. DEAB was used to establish the baseline fluorescence of these cells and to define the ALDEFLUOR-positive region.
Incubation of cells with ALDEFLUOR substrate in the absence of DEAB induces a shift in BAAA fluorescence, defining the ALDEFLUOR-positive population. (d).
Flow cytometric analysis of ESA+/PROCR+ population in MDA-MB-231 stable cells
(KLF4-N and Control).
Supplementary Figure 4. Kenpaullone treatment inhibited the promoter activities of
KLF4 and IAP. (a). Transcriptional activities of KLF4 was detected by a dual luciferase reporter assay after Kenpaullone treatment. MCF-7 cells were seeded in
12-well plates and cotransfection experiments were performed with 800ng of pGL3-KLF4 promoter and 50ng of pTK for 24 h. Cells were incubated with
Kenpaullone (5 µM) for an indicated time points (0, 1, 2, and 4 h) and the luciferase activity was measured using the dual-luciferase reagent assay kit (Promega). Activity of the firefly luciferase was normalized to that of the renilla luciferase and expressed as relative luciferase unit (RLU). * P < 0.05 vs Ken- group. (b). Similar to (a) except the IAP promoter was used.
Supplementary Figure 5. Inhibition of the PKA pathway increased KLF4 gene expression. MCF7 cells were treated with 5µM Kenpaullone, PI3K/Akt inhibitor
(50nM Wortmannin), ERK inhibitor (10µM PD98059), and PKA inhibitor 10µM H89.
24 hours after treatment, total RNAs were extracted and real-time PCR was
performed to detect KLF4 expression. * P < 0.05 vs DMSO-treated group.
Supplementary Figure 6. Reduction of the CD44+/CD24- population by Kenpaullone was mediated by KLF4. MCF-7 cells (siCon and siKLF4) were treated with 5 µM
Kenpaullone for 7 days followed by flow cytometric analysis as described in
Materials and Methods.
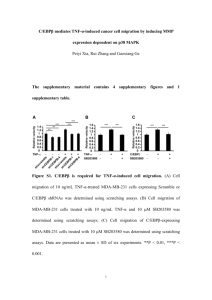
Supplementary Figure 7. Kenpaullone treatment had minimal effect on migration of
KLF4 knockdown cells. MDA-MB-231 (siCon and siKLF4) cells were seeded into
6-well plates and treated with 5 µM Kenpaullone. Wound closure rate was evaluated
12 h after Kenpaullone treatment.