Uniprot & Interpro tutorial - European Bioinformatics Institute
advertisement
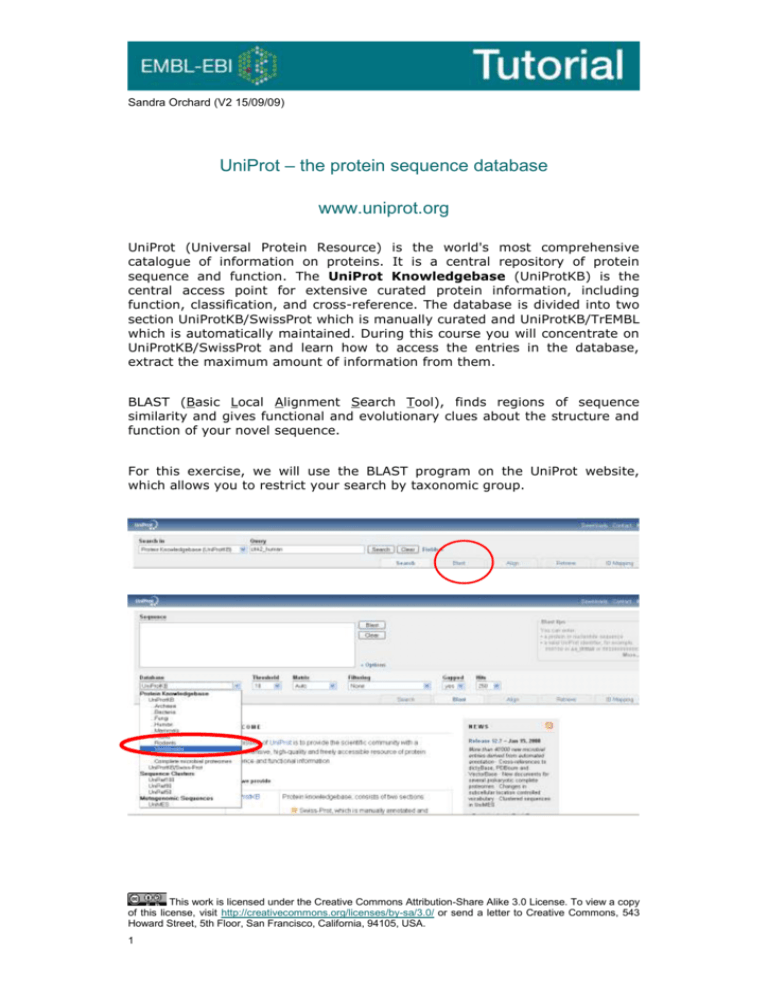
Sandra Orchard (V2 15/09/09) UniProt – the protein sequence database www.uniprot.org UniProt (Universal Protein Resource) is the world's most comprehensive catalogue of information on proteins. It is a central repository of protein sequence and function. The UniProt Knowledgebase (UniProtKB) is the central access point for extensive curated protein information, including function, classification, and cross-reference. The database is divided into two section UniProtKB/SwissProt which is manually curated and UniProtKB/TrEMBL which is automatically maintained. During this course you will concentrate on UniProtKB/SwissProt and learn how to access the entries in the database, extract the maximum amount of information from them. BLAST (Basic Local Alignment Search Tool), finds regions of sequence similarity and gives functional and evolutionary clues about the structure and function of your novel sequence. For this exercise, we will use the BLAST program on the UniProt website, which allows you to restrict your search by taxonomic group. This work is licensed under the Creative Commons Attribution-Share Alike 3.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/ or send a letter to Creative Commons, 543 Howard Street, 5th Floor, San Francisco, California, 94105, USA. 1 Exercise 1 – Exploring UniProtKB Go to the BLAST tool on the UniProt.org website. Paste in the Mystery protein sequence below. Restrict the search to Vertebrates. Mystery Protein sequence MSRESDVEAQQSHGSSACSQPHGSVTQSQGSSSQSQGISSSSTSTMPNSSQSSHSSSGTL SSLETVSTQELYSIPEDQEPEDQEPEEPTPAPWARLWALQDGFANLECVNDNYWFGRDKS CEYCFDEPLLKRTDKYRTYSKKHFRIFREVGPKNSYIAYIEDHSGNGTFVNTELVGKGKR RPLNNNSEIALSLSRNKVFVFFDLTVDDQSVYPKALRDEYIMSKTLGSGACGEVKLAFER KTCKKVAIKIISKRKFAIGSAREADPALNVETEIEILKKLNHPCIIKIKNFFDAEDYYIV LELMEGGELFDKVVGNKRLKEATCKLYFYQMLLAVQYLHENGIIHRDLKPENVLLSSQEE DCLIKITDFGHSKILGETSLMRTLCGTPTYLAPEVLVSVGTAGYNRAVDCWSLGVILFIC LSGYPPFSEHRTQVSLKDQITSGKYNFIPEVWAEVSEKALDLVKKLLVVDPKARFTTEEA LRHPWLQDEDMKRKFQDLLSEENESTALPQVLAQPSTSRKRPREGEAEGAETTKRPAVCA AVL The resulting scores should suggest a 100% match to CHK2_HUMAN (O96017). You should also note a match to the isoforms of CHK2 – each isoform is treated as a separate transcript, information which is often lost when using other protein sequence databases. Click on the top green bar in the Local Alignment column to display the aligned sequences. The UniProt Consortium is comprised of the European Bioinformatics Institute, the Swiss Institute of Bioinformatics and the Protein Information Resource. The UniProt consortium aims to support biological research by maintaining a high quality database that serves as a stable, comprehensive, fully classified, richly and accurately annotated protein sequence knowledgebase, with extensive cross-references and querying interfaces freely accessible to the scientific community. All data stored in UniProt can be downloaded from the Download Centre at http://www.ebi.uniprot.org/database/download.shtml Return to the search page and open O96017/CHK2_HUMAN. Click on the EC number link under protein names. Q 1. What is the reaction catalysed by this enzyme? Return to the UniProtKB/Swiss-Prot entry and click on the taxonomic identifier. 2 The NEWT database is a compilation of the information within the NCBI Taxonomy database together with proteins found in the Swiss-Prot and TrEMBL section of UniProtKB. It is maintained by the Swiss-Prot group in Switzerland. For each species, NEWT displays the following taxonomy data: Swiss-Prot scientific name, Swiss-Prot common name and Swiss-Prot synonym, lineage, number of protein sequence entries in Swiss-Prot and TrEMBL. The NEWT data is available from the European Bioinformatics Institute. Q 2. In UniProtKB how many proteins are there from Homo sapiens neanderthalensis? (Hint – Use the taxonomic hierarchy.) Return to the entry and scroll down. Q 3. How many other species also have proteins which are members of the CHK2 protein kinase subfamily? Return to the entry and scroll to Binary Interation. Q 4. How many evidences are there for the interaction of CHK2 with PLK1. Click on the hyperlink to IntAct to see which publication these evidences come from. Alternative Products in UniProtKB Return to the entry and go to the Alternative Products section. Chek2 is a protein for which multiple (12) isoforms have been identified. All of these are mapped within UniProtKB, and given stable identifiers. Press the button “Align” to see how all the isoforms differ. Return to the Alternative Products section. Q 5. What functional characteristic has the alternative splicing has conferred to isoform 12 (and other isoforms)? 3 To see the location and length of the missing region in isoform 12 go the UniProt entry (click O96017) and look in the Sequence annotation section under Natural variations. Q 6. Using the information in the table, explain why isoform 12 is catalytically inactive? (Hint – where’s the active site.) Q 7. Where in the cell is isoform 12 to be found? (Hint – look in Comments –> Subcellular location) Q 8. What is the length of isoform 12? Adding information using the InterPro Database At the bottom of the entry, are all the database cross-references for this protein. Look for those contained within the InterPro database. InterPro is an integrated documentation resource for protein families, domains and sites. InterPro combines a number of databases (referred to as member databases) that use different methodologies and a varying degree of biological information on well-characterised proteins to derive protein signatures. By uniting the member databases, InterPro capitalizes on their individual strengths, producing a powerful integrated diagnostic tool Signatures describing the same protein family, domain repeat or site are grouped into unique InterPro entries. Each combined InterPro entry has a unique accession number, an abstract describing the features of proteins associated with the entry and literature references and has links to the relevant member database(s). All UniProtKB protein sequences that have matches to a particular InterPro entry are listed in the Match Table associated with that entry. There are also links to the InterPro graphical views. The graphical views, which can be sorted by UniProtKB accession number, structure or taxonomy, show the position of the signatures on the protein, mousing over the signature brings up a pop-box, giving the accession, name and position. InterPro graphically represents the location of a protein domain and information pertaining to the origin of that domain and the proteins that contain it. Families are also defined and may contain several InterPro domains which are often, but not always, in the same order. InterPro and InterProScan are accessible for interactive use over the EBI web server (www/ebi.ac.uk/interpro), they are distributed as stand-alone copies by anonymous ftp. InterPro entries can be linked to one another through PARENT/CHILD and CONTAINS/FOUND IN relationships. PARENT/CHILD relationships indicate 4 superfamily/family/subfamily relationships, as well as domain hierarchies, where sequences can be subdivided into more specific sub-sets. CONTAINS/FOUND IN relationships apply to domains, repeats and sites within families, and are used to describe the composition of protein sequences. InterPro entries that belong to a UniProtKB entry can be found in Crossreferences -> Family and domain databases Moving back and forwards between the entry and the InterPro domains and active sites which it contains, answer the following questions Exercise 2 – Searching InterPro using a UniProt identifier Open the development version of the InterPro homepage (InterPro beta) in a web browser (http://wwwdev.ebi.ac.uk/interpro/). Using the “Text” Search box mid-way down the page, type in the UniProtKB accession ‘O15075’ (without the quotes. That’s a letter O at the start and a zero in the middle). Click on the purple “Search” button. You should now have a page describing the signature matches for this protein. (the protein view): Question 1: Looking at the InterPro protein view for O15075, how many InterPro entries (not individual signatures) match the query protein sequence? Question 2: How many domains is the protein divided up into? On the protein page for O15075, click on “Detailed results” in the left hand side menu. This will show you all of the protein signatures within InterPro that were used to create the information in the preceding page. Question 3: How many member database signatures contribute to InterPro entry IPR003533? Hint: You can see the contributing member database signatures to InterPro entry IPR003533 in the “Detailed results” view or alternatively click on the link to IPR003533, which will take you to the entry page for that domain. Go back to the “Overview” and scroll down to the “Structural features” just below the view of unintegrated signatures. Under the “Structural Features” heading you will find the PDB structure. Its length indicates the region of the protein for which the structure is known. You will also see bars representing a CATH database match and a SCOP database match, both of which are structural classification databases that break down the PDB structures for the protein into their constituent domains. 5 Question 4: What region is covered by the PDB structure (ie which domain)? Hint: Compare it to entry IPR003533. Not all of the protein has been structurally characterised, shown by the fact that only a small region of this protein is covered by the PDB match. To help address this problem, there are homology models from both ModBase and Swiss-Model found under the “Structural Predictions” section. These are models based on aligning our protein with its closest homologue whose structure has been determined. (Note: these are predictive models that provide a ‘best guess’ at the remaining structure). Question 5: Why does IPR003533 have two domain hits compared to the single domain the PDB structure? Note the structural view at the top right of the page and click on the ‘Go’ purple bottom. It will bring you to the PDB page, where you can visualise the 3-D structure by clicking on Jmol (it will pop up in a new window). Exercise 3 – Exploring InterPro entries General annotation Still on the protein page for O15075, look at the match to the entry IPR000719. Notice that there are other domain entries that cover approximately the same sequence position. Click on the hyperlink to IPR000719 Question 1: What is the name of this domain? Now look at the “Contributing signatures” section. Question 2: Which signatures make up this entry? This section lists the signatures in an entry, the database they come from, and the number of proteins they match. Relationships Question 3: What “Child” entries is IPR000719 subdivided into? InterPro links related signatures through Parent/Child relationships which indicate domain/family hierarchies. Child entries subdivide IPR000719 into more closely related subgroups. Question 4: What is the name of the “Parent” of IPR000719? In this case, the parent entry represents domains with a structural fold homologous to that of the protein kinase domain (even if they have no enzyme activity), whereas IPR000719 represents a more specific form of the domain that has catalytic protein kinase activity. GO (Gene Ontology) terms Scroll down to the “GO terms annotation” section. 6 Question 5: What GO terms are provided for this entry? InterPro provides its own mappings to GO terms based on the curated UniProt/Swiss-Prot proteins matching an entry. These are useful for the annotation of TrEMBL proteins that do not otherwise have GO terms associated with them. (Note: Next to the Go terms you can find a link to other entries in InterPro that share that Go term. This is of value if you are interested in searching for InterPro entries that match proteins with a specific function or those involved in a specific process). Now look at the information in the side menu for entry Question 6: How many proteins are matched by IPR000719? Structures Now click on the "Structures" link on the left hand side menu. InterPro provides a list of all the PDB entries associated with an entry. There are also structural links to SCOP and CATH at the bottom of the page, which provide structural classifications of the proteins that match this entry. Scroll to the bottom of the page and follow the “SCOP d.144.1.7” link to the SCOP database to find out the structural classification of this domain. Question 7: What type of structure does the protein kinase-like fold consists of? Hint: look at the information under “Fold” in the “Linage” section. Taxonomy Click on the browser back button till you are in the InterPro page again, then click on the "Species" link on the left hand side of the InterPro entry page for IPR000719. You may explore the taxonomic spread by expanding the table. InterPro divides all the protein hits in an entry by their taxonomy. Question 8: How wide a taxonomic coverage do proteins containing a protein kinase domain have? 7 Searching UniProtKB Return to the search engine at the top of the entry page and click on Fields Use the fields to search on IPR000719 AND Organism=human to see how many protein kinases you can find. Q 11. How many of those are in Swiss-Prot (i.e. reviewed records)? Q 12. Furthermore, how many of those have experimental evidence of being associated with the cell membrane? (As well the subcellular location fields in the Comments section other fields such keywords and gene ontology also have subcellular location information. However, in the UniProt search facility only the subcellular location fields allows the evidence to taken in to account.) 8 Further Info Sequence Searching For more sequence searching tools visit http://www.ebi.ac.uk/Tools/similarity.html. There are two main programs that implement BLAST searches; WU-BLAST 2.0 and NCBI BLAST2. They are distinctly different software packages, although they have a common lineage for some portions of their code, so the two packages do their work differently and obtain different results and offer different features. You can also check for vector contamination with Blast2 EVEC. Fasta can be very specific when identifying long regions of low similarity especially for highly diverged sequences. You can also conduct sequence similarity searching against complete proteome or genome databases using the Fasta programs. MPsrch – Smith and Waterman algorithm, capable of identifying hits in cases where Blast and Fasta fail and also reports fewer false-positive hits. Further Reading The UniProt Consortium “The Universal Protein Resource (UniProt).” Nucleic Acids Res. (2008) 35:D193-197 Leinonen R, Nardone F, Zhu W, Apweiler R “UniSave: the UniProtKB sequence/annotation version database.” Bioinformatics (2006) 22:12841285 Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH “UniRef: comprehensive and non-redundant UniProt reference clusters.” Bioinformatics (2007) 23:1282-8 Mulder NJ et al “New developments in the InterPro database.” Nucleic Acids Res. (2007) 35:D224-8 Kerrien S et al “IntAct--open source resource for molecular interaction data.” Nucleic Acids Res. (2007)35:D561-565 Exercise 1 – UniProt 9 Q 1. What is the reaction catalysed by this enzyme? ATP + a protein = ADP + a phosphoprotein Q 2. In UniProtKB how many proteins are there from Homo sapiens neanderthalensis? 23 Q 3. How many other species also have proteins which are members of the CHK2 protein kinase subfamily? 13 proteins in 6 species Q4 How many evidences are there for the interaction of CHK2 with PLK1. Click on the hyperlink to IntAct to see which publication these evidences come from. 6 (Jan 2012) – PMID: 12493754 Q 5. What functional characteristic has the alternative splicing has conferred to isoform 12 (and other isoforms)? Inactive Q 6. Using the information in the table, explain why isoform 12 is catalytically inactive? Alternative splicing removes the active site Q 7. Where in the cell is isoform 12 to be found? Nucleus Q 8 What is the length of isoform 12? 514 Exercise 2 - Searching InterPro using a UniProt identifier. Question 1: Looking at the InterPro protein view for O15075, how many InterPro entries (not individual signatures) match the query protein sequence? Answer 1: Seven (six domain/sites entries plus the family membership entry) Question 2: How many domains is the protein divided up into? Answer 2: Three (one of them is found repeated in the protein) Question 3: How many member database signatures contribute to InterPro entry IPR003533? 10 Answer 3: Five Question 4: What region is covered by the PDB structure (ie which domain)? Answer 4: The first doublecortin domain of the two represented by InterPro entry IPR003533. Question 5: Why does IPR003533 have two domain hits compared to the single domain the PDB structure? Answer 5: IPR003533 predicts the presence of two doublecortin domains, but only the area corresponding to the first one has been structurally characterised and therefore appears in the PBD structure. Exercise 3 - Exploring InterPro entries Question 1: What is the name of this domain? Answer 1: Protein kinase, catalytic domain Question 2: Which signatures make up this entry? Answer 2: Two signatures make up this entry (PF00069 and PS50011) Question 3: What “Child” entries are IPR000719 subdivided into? Answer 3: Two: Serine-threonine/tyrosine protein kinase catalytic domain and Serinethreonine/dual-specificity protein kinase catalytic domain Question 4: What is the name of the “Parent” of IPR000719? Answer 4: Protein kinase-like domain Question 5: What GO terms are provided for this entry? Answer 5: GO:0006468 (protein phosphorylation), GO:0004672 (protein kinase activity), GO:0005524 (ATP binding) Question 6: How many proteins are matched by entry IPR000719? Answer 6: 111054 proteins are matched Question 7: What type of structure does the protein kinase-like fold consists of? 11 Answer 7: Consists of two alpha+beta domains, and the C-terminal domain is mostly alpha helical. Question 8: How wide a taxonomic coverage do proteins containing a protein kinase domain have? Answer 8: It is widely spread, being found in eukaryota, bacteria, archaea and viruses. Exercise 3- Analysing sequences using InterProScan Question 1: Based on the InterPro matches, what domains is the protein predicted to possess? What family is it predicted to belong to? Answer 1: A six-hairpin glycosidase-like domain (IPR008928) and belongs to the Lanthionine synthetase C-like family (IPR007822), subfamily LanC-like eukaryotic (IPR20464). Question 2: What do your BLAST results suggest your protein to be? Answer 2: G protein coupled receptor Question 3: Are the results consistent with those returned by InterProScan? Is there anything in the InterPro annotation that might explain any discrepancies? Answer 3: InterPro entry IPR007822 explains that due to its structure some authors considered LanC-1 and other members of the family to be novel G protein-coupled receptors, but this claim has since being refuted. Nevertheless, many sequences in the protein databases were submitted as G protein-coupled receptors and still carry this description. This creates some confusion when doing a BLAST search. 12