Supplementary material TRFLP profile of sea water and coral mucus
advertisement

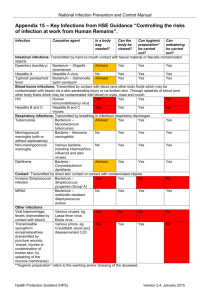
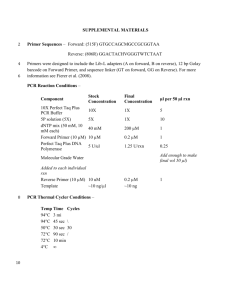
Supplementary material TRFLP profile of sea water and coral mucus Terminal Restriction Fragment Length Polymorphism (TRFLP) analysis was done for the same mucus metagenome used in this study and the surrounding sea water metagenome which showed that the bacterial communities associated with the mucus and the surrounding sea water were entirely different. For TRFLP analysis, the universal 27F forward 16S primer was labeled at the 5’ end with phosphoramidite fluorochrome 6-carboxyfluorescein (6-FAM). TRFLP was done according to Klaus et al; 2007 where the amplicons labeled with 6-FAM were digested with the restriction enzyme MspI. The Gene scan analysis was done at Macrogen, Korea. Peak scanner software was used to generate tables of peak area and fragment size data for each sample. Peak area and fragment size of Sea water -MspI Peak area and fragment size of coral mucus -MspI TRFLP analysis was done using T-RFLP FRAGSORT. T-RFLP FRAGSORT is a computer program that compares the TRFs obtained from samples processed with widely used primers (8F, 907R, 27F, 226F, 1111R or EF4) and restriction enzymes (Msp1, Hha1, Rsa1, HaeIII, and Bfa1) to TRFs from corresponding in silico amplification and digestions of RDP II alignments (version 8.0). FRAGSORT input consists of one, two or three data files containing the fragment sizes and normalized peak areas (or heights) from restriction digestions of environmental 1 DNA amplified with a fluorescent primer. The input file can be in text, csv or genescan format. The program outputs organisms that match 1, 2 or 3 T-RFLP fingerprints. The program compiles a list of organisms matching the TRFs from each restriction enzyme, then determines which organisms match 2 or 3 of the different fragments. The program output is a list of microorganisms, TRF sizes, and normalized peak areas that correlate with multiple experimentally generated T-RFLP profiles. MspI Profile of Sea water MspI 1.2 Normalized TRF Peak Area (%) 1 0.8 0.6 0.4 0.2 10 9 13 0 13 5 15 5 15 9 16 0 16 0 16 0 16 0 16 0 16 0 16 0 16 1 16 1 16 6 20 2 27 7 33 0 48 4 84 72 65 0 TRF Fragm ent Size (bp) MspI Profile of Coral mucus Msp I 1.4 1 0.8 0.6 0.4 0.2 TRF Fragment Size (bp) 2 486 485 485 485 484 448 430 291 184 158 157 157 157 157 149 141 127 126 126 126 126 125 125 125 125 125 122 122 122 88 122 88 88 88 88 87 87 80 71 71 70 0 63 Normalized TRF Peak Area (%) 1.2 Supplementary Table S1. Representative output list of microorganisms present in the surrounding sea water Sea water 1 Candidatus Tremblaya princeps. 2 Candidatus Tremblaya princeps. 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 uncultured proteobacterium a2b027 1-2 cm layer of core A. uncultured bacterium mle1-31. uncultured bacterium DR9IPCB16SCT4. Desulfurella acetivorans (T) DSM 5264. Syntrophus sp.. Syntrophus buswellii (T) DSM 2612. uncultured bacterium SHA-51. uncultured bacterium IndB4-24. Lactococcus lactis lactis K338. Actinocorallia aurantiaca (T) JCM8201. Actinocorallia glomerata IMSNU 22179T. Actinocorallia longicatena IMSNU 22180T. Mycobacterium sp. IP20010664. Mycobacterium sp. Mycobacterium savoniae type strain: E533. Mycobacterium smegmatis mc2 155. Thermotoga lettingae (T) TMO. Propionibacterium freudenreichii JS53. Corynebacterium sp. 61720. uncultured bacterium O-F-5. Corynebacterium sp. CIP102857. Corynebacterium sp. CIP102645. Corynebacterium sp. CIP102622. Corynebacterium sp. CIP102590. Corynebacterium sp. CIP102346. Corynebacterium sp. CIP102211. Corynebacterium sp. CIP102124. Corynebacterium sp. CIP102076. Corynebacterium sp. CIP101775. Corynebacterium sp. CIP107291. Corynebacterium sp. CIP107067. Corynebacterium segmentosum CIP107068, 34 (CCUG37878). 3 35 36 37 38 39 40 41 Corynebacterium macginleyi CIP104099T, (ATCC51787T). Corynebacterium accolens CIP104783T, (ATCC49724T). Corynebacterium fastidiosum CIP103808. Corynebacterium pseudogenitalium CIP106714, (ATCC33035), uncultured Corynebacterium sp. ACTINO9B. Gordonia nitida J-1. Gordonia alkalivorans. Representative output list of microorganisms present in the coral mucus 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Coral Mucus bacterium Te32A. 22 uncultured bacterium SOB-13. bacterium Te45R. 23 uncultured proteobacterium OCS7. bacterium Te58R. 24 uncultured bacterium O-CF-10. bacterium Te47R. 25 uncultured bacterium C-F-17. uncultured bacterium D137. 26 bacterium H15. bacterium Te37R. 27 uncultured bacterium FB33-34. bacterium Te49A. 28 uncultured bacterium FB47-42. Enterococcus sp. GvF410. 29 uncultured bacterium FB49-2b. Enterococcus faecium E 26. 30 uncultured bacterium 015B-F04. uncultured isopod gut bacterium RKPsAG. 31 uncultured bacterium 015C-F06. Enterococcus faecalis UK873 32 Zoogloea ramigera ATCC 25935. bacterium Te6A. 33 uncultured bacterium 4f39. bacterium Te50A. 34 uncultured sludge bacterium A35. bacterium Te95A. 35 uncultured bacterium SHA-58. 15 Pseudoalteromonas bacterium . 36 gamma proteobacterium HTCC230. 16 Rhodothermus marinus 10. 37 Rhodothermus marinus (T) R-10 17 (DSM 4252). 38 gamma proteobacterium HTCC234. uncultured Sphingobacteriales bacterium SF62. 18 uncultured bacterium IheB3-31. 39 uncultured Sphingobacteriales 19 bacterium SF63. 41 uncultured bacterium KD8-91. 20 unidentified bacterium JTB326. 42 uncultured bacterium FukuN9. 21 Psychrobacter sp (T). 43 Uncultured rumen bacterium F24-F11. uncultured bacterium KD8-112. 4