transcript
advertisement

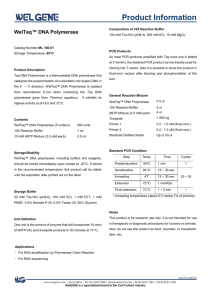
New polymerases for old DNA Phil Holliger MRC LMB Slide1: Top left In 79 A.D., Mount Vesuvius erupted and buried two towns. One of these was Pompeii, the other was Herculaneum, a seaside resort for wealthy Romans. Top right Among the villas uncovered by excavation was the summer residence of Julius Caesar's father-in-law, Lucius Calpurnius Piso. The picture shows a reconstruction of the villa forming part of the Getty museum in LA. Bottom left Inside the villa a large number of what appeared to be sticks of charcoal were discovered, some of them bundled together. Bottom Middle These turned out to be ancient papyrus scrolls carbonized by the volcanic heat. The “Viilla dei Papyrii” as it became known, contained the only known library of antiquity. Bottom right While some part of the text remained legible, large parts of the scrolls were initially uninterpretable. However, the information encoded in this ancient library was not lost but had to wait for better technology. Today, multi-spectral imaging promises a breakthrough in deciphering the fragile scrolls. Slide 2: By analogy, many specimens paleontological, archaeological, or forensic interest contain a wealth of information written in their DNA. Ancient DNA sequences have been isolated from a wide variety of sources 1 and have provided information about human migration, animal and crop domestication and the genetic relationship between modern Homo sapiens and its closest extinct relative H. neandertalensis.(left panel) Such ancient DNA can provide a window to the past but even under optimal burial conditions, DNA damage is progressive, as the multitude of DNA repair pathways, which maintain the integrity of the genome in living organisms no longer function. The right panel shows a summary of the multitude of types of damamge found in ancient DNA. This damage either limits the length of continuous sequence that can be recovered or renders even well-preserved specimens unproductive despite the demonstrable presence of DNA (by hybridization) We reasoned that genetic information encoded in such samples may not be lost but simply inaccessible due to the fact that the DNA polymerases commonly used for PCR stall at sites of damage. Polymerases capable of replicating across DNA damage should therefore be able to allow the deciphering of previously unreadable ancient DNA sequences just like modern imaging is helping decipher the burnt scrolls of Herculaneum. However, no such polymerases suitable for ancient DNA recovery existed in nature. We therefore turned to evolution to generate such polymerases in the laboratory using nature’s tricks. Slide 3 Darwinian evolution can be applied not just to organisms but to molecules too. Thus, molecular properties can be improved by iterative cycles of mutation, selection and amplification. Slide 4 We devised a strategy for the selection of polymerase function, which we call “compartmentalized self-replication” or CSR. CSR is based on a simple feedback loop, whereby a polymerase replicates its own encoding gene. Compartmentalization serves to isolate individual self-replication reactions from each other. Compartmentalization is a crucial aspect of life. All living organisms are made from cells, which encase the genome and the proteins it encodes within a lipid membrane. We use a different approach to nature. In our case, polymerase genes and the polymerase they encode are encapsulated in water droplets dispersed in an oil phase, i.e. a water-in-oil emulsion. These “artificial cells” ensure genotype-phenotype linkage, i.e. they ensure that a polymerase only replicates only their own encoding gene and no other. In such a system adaptive gains by a polymerase directly (and proportionally) translate into more “offspring”. In other words, if a polymerase (purple spheres) is well-adapted to the selection conditions, it can replicate its own encoding gene and produce many copies of itself, i.e. “offspring”. However, if a polymerase (yellow hexagon) is poorly adapted, it cannot self-replicate and its gene will disappear from the gene pool. Slide 5 Left panel When random mutagenesis of the polymerase genes proved unproductive we turned to a method called molecular breeding, whereby the homologous genes from different organisms (orthologues) are recombined to yield a library of chimeras comprising segments of the different orthologues. Molecular breeding samples only functional diversity and therefore a large number of chimeras are active. We recombined three thermophilic polymerase (DNA pol I) genes from the genus Thermus: Taq polymerase from (Thermus aquaticus), the standard polymerase used for PCR amplification of ancient DNA, Tth (T. thermophilus) and Tfl (T. flavus)) to create a polymerase library for selection. Right panel This shows a three dimensional model of Taq polymerase. Residues deriving from Tth or Tfl that we find in our selected polymerases are shown in different shades of blue. The darker the blue the more often they occur. This just to illustrate how the offspring of an evolution experiment can comprise a patchwork of elements of the parental genes. Slide 6 Evolving polymerases, which combine the processivity and selectivity required for PCR amplification with a high tolerance for template damage is challenging. Furthermore, damage tolerance should be generic as detailed information about the forms of DNA damage in ancient samples is lacking (and damage may vary depending on burial conditions). Top Many lesions (red X) abrogate base-pairing and yield distorted, non-cognate 3’ structures, similar to mismatches. While natural polymerases readily extend of matched primer terminus (ending in a cognate GC base-pair), they stall at mismatches or sites of damage (G.X mispair). Extension is significantly slowed down not just at the 3’ end but up to four bases downstream (highlighted in red). In order to maximize tolerance to such distorted primer-template structures, we decided to select for polymerases capable of extending a primer 3’ terminus preceded by up to four mismatched bases. Bottom left This shows the selection scheme. Two independent aqueous compartments of the water-in-oil emulsion are shown. Polymerases (such as Pol1 (left compartment)) that are capable of utilizing quadruple mismatch primers (AGGGAGGG, GGTGGGTG) to replicate their one encoding gene (pol1) produce “offspring” i.e. increase their copy number in the post-selection population, while polymerases like Pol2 (right compartment) that are unable to utilize quadruple mismatch primers disappear from the gene pool Bottom right After three rounds of CSR selection, we recovered a diverse set of polymerases with novel properties including the generic ability to utilize single, double and quadruple mismatches (as seen in this picture for the polymerases called 3D1). They could also process unusual DNA structures and bypass template lesions such as abasic sites or hydantoins, as will be shown in the following two slides. Slide 7 Top left We examined primer extension reactions using a radiolabelled primer and resolved products on a polyacrylamide gel. We examined extension of three different quadruple mismatches: (M1: GGTGGGTG, M2: AGGGAGGG)(used for selection), and the unrelated (M3: TTTTTTTT) and compared it to extension of matched primer (M0). While Taq was unable to extend any of the mismatches , theselected polymerases 3A10 and 3D1 yield extension products with M1-3 but extension products are predominately shorter than M0. Top right and bottom Possible primer template configurations and expected main product lengths (N+1) are illustrated. Matched primer-template sequences (M0) at primer 3’ end are shown in blue, mismatched and misaligned structures are shown in red. Slide 8 Again we measured polymerase activity by examining the ability to extend a radiolabelled primer. Top left On an undamaged template all three polymerases Taq and the selected polymerases 3A10, 3D1 display approximately the same activity. To the left, the chemical structure of the undamaged base T is shown. Top right This template contains an abasic site at the + 1 position (marked by a red AP). To the right, the chemical structure of an abasic site is shown. Abasic sites are among the most frequent forms of DNA damage and are generated by spontaneous depurination or depyrimidination and as the end product of a number of oxidation-induced DNA damage pathways. As can be seen, while Taq polymerase can insert a nucleotide opposite the abasic site (see arrow), it cannot bypass and remains stuck in the +1 position. In contrast, the selected polymerases 3A10(and to a lesser extent 3D1) can efficiently bypass the site of damage, inserting mostly an A opposite the absic site. Bottom left and right These template contains hydantoins at the + 1 position (marked by a red 5H, 5M). Hydantoins are oxidative degradation products of the pyrimidine bases. 5-hydroxy-hydantoin derives from C and 5-methyl-5-hydroxy-hydantoin derives from T. High levels of hydantoins have been found in some ancient samples and associated with PCR failure. Their chemical structures are shown to either side of the gel pictures. These lesions show the same general picture. Taq polymerase can insert a nucleotide opposite the hydantoins (see arrow), but bypasses them poorly 5hydroxy-hydantoin very poorly. The selected polymerases 3A10 also have problems in bypassing the lesions but 3A10 is significantly better than Taq on either lesion, inserting mostly an A in both cases. Slide 9 The ability of 3A10 to bypass template damage is reflected in PCR amplification. This shows a gel picture of the PCR amplification of DNA containing 2 abasic sites. As can be seen while all natural polymerases such as Taq but also Tth and Tfl fail to yield a PCR amplification product, 3A10 yields a strong amplification band. The other weaker bands are from other selected polymerases. Slide 10 Left panel The ability of selected polymerases to efficiently bypass template lesions in PCR encouraged us test their activity for the recovery of ancient DNA. We performed subsequent experiments using a blend of Taq with the most promising selected polymerases (3A10, 3D1 and others) (rather than testing individual combinations) in order to minimize wastage of precious ancient samples and maximize the chances of success. We first performed 56 PCR amplifications at limiting dilutions of ancient DNA (aDNA) derived from a 47,000 year-old cave bear (Ursus spelaeus) bone and scored successful amplifications for blend and Taq alone. We found that the blend yielded amplification products at between 2 - 5-fold lower concentrations of aDNA than Taq and indeed did yield amplification products at DNA concentrations, where Taq no longer generated any Right panel Normalizing PCR activity on a dilution series of “modern” DNA showed that this was not due to higher PCR efficiency of the blend. On the contrary, Taq appeared to be more than an order of magnitude more active at low template concentrations (of “modern” DNA), suggesting that the blend requires more template than Taq to produce an equivalent PCR signal. The increased template DNA requirement of the blend suggests that the increased ability of the blend to amplify ancient DNA represents an underestimate of the blend’s potential. Moreover, it implies that the blend can tap into a pool of DNA molecules that are inaccessible to Taq, presumably because they are damaged. Slide 11 To stringently exclude sample heterogeneity and stochastic variation as the source of the above effect, we performed a further 608 independent PCR amplifications from two different samples of cave bear bone (~47,000 and ~60,000 years-old respectively), and scored the number of PCR amplicons at limiting dilution. The blend yielded a larger number of amplicons (8-150%) than Taq in all but one experiment, confirming previous results. In conclusion, molecular breeding and directed evolution by CSR have allowed the isolation of polymerases, which enhance the recovery of genetic material from Pleistocene specimens, presumably due to their ability to amplify damaged DNA. Ice age genomics is upon us. Largely, thanks to novel sequencing methods, such as the Roche /454 sequencer, which also utilize emulsion PCR, Polymerases such as those described here should benefit the recovery of ancient DNA and may speed up sequencing as they are pre-adapted to emulsion PCR. Polymerases capable of amplifying damaged DNA may also reduce bias towards modern DNA contamination and enable novel applications in palaeobiology, molecular archaeology and historic and forensic medicine. The novel polymerases described here are really just a step in a direction, but they show that we can use evolution to improve our molecular tools. Further improvements should be within reach and hopefully will render ever more ancient sequences readable to us.