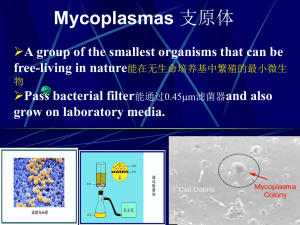
abstracts - Unisi.it
advertisement

Competence-Specific RNA Polymerase in Streptococcus pneumoniae: the Role of ComX. PING LUO AND DONALD A. MORRISON* Laboratory for Molecular Biology, Department of Biological Sciences, University of Illinois at Chicago, Chicago Illinois 60607 Natural competence in Streptococcus pneumoniae is regulated by a quorum-sensing system which acts through accumulation and sensing of a peptide pheromone (CSP) to control many competence-specific genes acting in DNA uptake, processing, and integration. The period of competence induced by CSP lasts only 20-30 minutes. The recently identified regulator ComX is encoded by duplicate genes and is required for the induction of many competence-specific genes in response to CSP. The shutoff of CSPinduced gene expression also depends on ComX. As genes regulated by ComX share an unusual consensus sequence (TACGAATA) at their promoter region, it is thought that this modulator might act as a transient alternative sigma factor to direct RNA polymerase to specific promoters. To test this hypothesis, ComX was purified from an E. coli overexpression strain and core RNA polymerase was purified from comX-deficient S. pneumoniae. Using PCR products with representative promoter regions as templates, it was shown that the reconstituted holoenzyme could transcribe competence-specific genes like ssbB, cinA, cglA, and celA, while holoenzyme prepared from non-competent cells did not. Western blotting using a specific Anti-ComX antibody showed that ComX existed transiently after CSP treatment, with a maximum at 15 min for a period of 20 min. Finally, Western blotting showed that ComX copurified with RNA polymerase from competent Streptococcus pneumoniae cells, indicating that ComX was associated with RNA polymerase in vivo during competence. At its maximal level, ComX was found in a molar ratio to RNA polymerase of approximately two. These data show that ComX is a sigma factor that enables transcription of competence-specific genes, and that it is an unstable protein whose synthesis begins during the lag period between the appearance of the CSP signal and the expression of some of the genes required for DNA uptake and processing during genetic transformation, and which does not persist after competence is shut off. INDUCTION OF NATURAL COMPETENCE IN STREPTOCOCCUS PNEUMONIAE TRIGGERS LYSIS AND DNA RELEASE FROM A SUBFRACTION OF THE CELL POPULATION. Hilde Steinmoen, Eivind Knutsen and Leiv Sigve Håvarstein Department of Chemistry and Biotechnology, Biotechnology building, Agricultural University of Norway, N-1432 Ås, Norway. Naturally competent bacteria have the ability to take up free DNA from the surrounding medium, and incorporate this DNA into their genomes by 1 homologous recombination. In naturally competent Streptococcus pneumoniae, and related viridans streptococci , the competent state is not a constitutive property, but is induced by a peptide pheromone through a quorum-sensing mechanism. Recent studies have shown that natural genetic transformation is an important mechanism for gene exchange between streptococci in nature. A prerequisite for effective gene exchange is the presence of streptococcal donor DNA in the environment. Despite decades of study of the transformation process we still do not know how this donor DNA is released from streptococcal cells to the external milieu. Traditionally it has been assumed that donor DNA originates from cells that die and fall apart from natural causes. In this study we show that induction of the competent state initiates release of DNA from a subfraction of the bacterial population, probably by cell lysis. The majority of the cells induced to competence take up DNA and act as recipients, while the rest release DNA and act as donors. These findings show that natural transformation in streptococci provides a natural mechanism for genetic recombination that in some aspects resembles sex in higher organisms. Spontaneous release of DNA and induction of competence in cultures of Streptococcus pneumoniae Miriam MOSCOSO and Jean-Pierre CLAVERYS. Laboratoire de Microbiologie et Génétique Moléculaire, UMR 5100 CNRS-Université Paul Sabatier. 118, route de Narbonne. 31062 Toulouse Cedex, France. Although it is generally well accepted that natural transformation plays a major role in the genetic plasticity of the human pathogen Streptococcus pneumoniae, the frequently asked question of the source of DNA in nature remains unanswered. Ottolenghi and Hotchkiss (J Exp Med, 1962; 116: 491-519) presented evidence suggesting that, in S. pneumoniae, maximum release of DNA with genetic transforming activity into the culture medium coincided with the development of competence. No evidence of concomitant cell disintegration or death was obtained in S. pneumoniae (Ottolenghi and Hotchkiss, ibid.). Although the published observations suggested the existence of a correlation between competence development and DNA release, no evidence for a direct link was provided. This prompted us to reinvestigate the phenomenon to establish whether release is causally related to competence, taking advantage of recent progress made in our understanding of the regulation of competence development and the availability of mutants affected in various steps in the process. We then investigated whether DNA release and the induction of competence are intimately connected by analyzing the kinetics of DNA release in mutants affected in the secretion of CSP (i.e. comAB) or in the expression of the competence control operon, comCDE. Results demonstrating that DNA release and competence development are causally related will be presented. As to the mechanism of competence-dependent DNA release, preliminary 2 experiments indicate that release is affected by the inactivation of lytA, which encodes the major pneumococcal autolytic enzyme. The Streptococcus pneumoniae CiaR/CiaH Two Component System and Genetic Competence: CiaR Target Fragments and cia-dependent Gene Expression Thorsten Mascher1, Dorothea Zähner1, Michelle Merai, Nadège Balmelle2, Antoine B. de Saizieu2, and Regine Hakenbeck1* 1 University of Kaiserslautern, Department of Microbiology, Paul Ehrlich Straße 23, D67663 Kaiserslautern, Germany; 2F. Hoffmann-La Roche Ltd, CH-4070 Basel, Switzerland The ciaR/ciaH system is one of thirteen two component systems of the human pathogen Streptococcus pneumoniae. Mutations in the histidine protein kinase CiaH confer increased resistance to beta-lactam antibiotics and interfere with the development of genetic competence. In order to identify the genes controlled by the cia system – the cia regulon – DNA fragments targeted by the response regulator CiaR were isolated from restricted chromosomal DNA using the solid phase DNA binding assay and analyzed by hybridization to an oligonucleotide microarray representing the S. pneumoniae genome. A set of thirteen chromosomal regions containing 17 CiaR target sites are proposed to represent the minimal cia regulon. The putative CiaR target loci included genes important for synthesis and modification of cell wall polymers and the pneumococcal htrA homologue. The data were complemented by analyzing the transcription profile of cia mutants representing the on and off state of the regulatory system. The transcript analysis confirmed the cia dependent expression of putative target loci, and documented that the entire competence regulon is also affected by the cia system. It is likely that the htrA locus which is located adjacent to the comCDE operon required for induction of genetic competence plays a key role in this regulatory network. Uptake of transforming DNA in Gram-positive bacteria: A view from Streptococcus pneumoniae Mathieu Bergé#, Miriam Moscoso #, Marc Prudhomme, Bernard Martin, and Jean-Pierre Claverys* Laboratoire de Microbiologie et Génétique Moléculaire, UMR 5100 CNRS-Université Paul Sabatier, 118 route de Narbonne, 31062 Toulouse Cedex, France. # These authors contributed equally to this study. 3 In a working model for uptake of transforming DNA based on evidence taken from both Bacillus subtilis and Streptococcus pneumoniae, the ComG proteins are proposed to form a structure that provides access of DNA to the ComEA receptor, through the peptidoglycan. DNA would then be delivered to the ComEC-ComFA transport complex. A DNA strand would be degraded by a nuclease while its complement would be pulled into the cell by ComFA, through an aqueous pore formed by ComEC. The nuclease is known only in S. pneumoniae, as EndA. We have examined the processing (i.e. binding, degradation and internalization) of DNA in S. pneumoniae strains lackings candidate uptake proteins. Mutants were generated by transposon insertions in endA, comEA/C, comFA/C, comGA and dprA. Processing of DNA was abolished only in a comGA mutant. As significant binding was measured in comEA mutants, we suggest the existence of two stages in binding: surface attachment (abolished in a comGA mutant) preceding deep binding (by ComEA). Degradation was abolished in comEA mutants indicating that, despite its membrane location, EndA cannot access donor DNA by itself. We propose that ComEA delivers DNA to EndA. Binding and degradation still occurred in comEC and comFA mutants. We conclude that recruitment of EndA readily occurs in the absence of ComEC or ComFA and that continuous action of EndA is not dependent on the pulling into the cell of single strands it produces. Finally, inactivation of dprA had no effect on internalization of DNA, indicating that DprA is required at a later stage in transformation. Competence for genetic transformation and virulence in Streptococcus pneumoniae : common regulatory steps and metabolic implication Trombe Marie-Claude EA 3036, IFR31 Centre Hospitalo Universitaire de Rangueil, Université Paul Sabatier, 31403 Toulouse France Tel ( ) 561322974, @mail : Trombe @CICT.fr Although genetic transformation has been discovered in vivo in a mouse model of infection, the rôle of competence development in virulence if any, is still an open question. Mutational analysis and genetic dissection allowed to provide evidence in favour of the implication of three two component signaling systems and of the metabolic functions NOX and REGR both in competence regulation and in virulence expression in mice. RegR is a global regulator of the lacI/GalR family which pleiotropically modulates virulence through its negative regulation of hyaluronidase and probably its positive effect on the expression of uncharacterized virulence factor(s). Nox is an NADH oxidase which reduces oxygen into water as it recycles NADH. Competence regulation studies and genetic dissection showed that both NOX and REGR act upstream of the signal transducing systems CiaRH and ComDE leading to optimal transformability of cultures growing aerobically. In addition, signaling via the two component signaling system MicAB which kinase MicB carries a PAS signature, is involved in competence regulation. 4 These data converge to show that common metabolic inputs and signaling pathways adjust the competence level in vitro as well as experimental virulence expression in response to a set of environmental parameters including oxygen. References - Echenique J., Trombe M.-C. 2001 Competence repression under oxygen limitation through the two component MicAB signal transducing system in Streptococcus pneumoniae, involvement of the PAS domain of MicB J. Bacteriol. 183 4599-4608 - Chapuy-Regaud S., Duthoit F., Malfroy-Mastrorillo L., Gourdon P., Lindley N.D., Trombe M.-C. 2001 Competence regulation by oxygen availability and by Nox is not related to specific adjustment of central metabolism in Streptococcus pneumoniae, J. Bacteriol. 183, 2957-2962 - Echenique J., Trombe M.-C. 2001 Competence modulation by Nox involves signal transduction in Streptococcus pneumoniae, J. Bacteriol. 183, 768-772 - Echenique J., Chapuy-Regaud S., Trombe M.-C., 2000 Oxygen regulation of competence in Streptococcus pneumoniae : involvement of ciaRH and comCDE, Molecular Microbiology 36, 688-696 - Auzat I., Chapuy-Regaud S., Dos Santos D., Le Thomas I, Le Bras G, Ogunniyi D, Garel J-R, Paton J., Trombe M-C, 1999 The NADH oxidase of Steptococcus pneumoniae, its role in competence and virulence, Molecular Microbiology 34, 1018-1028 - Chapuy-Regaud et al. Involvement of the lacI/GalR family transcriptional regulator RegR in virulence and competence of S. pneumoniae manuscript submitted MalR mutants: A new class of superepressor C. Nieto, M. García de laCoba, and M. Espinosa. Centro de Investigaciones Biológicas ,CSIC, Madrid, Spain The Streptococcus pneumoniae MalR protein negatively regulates transcription from two divergent operons malXCD and malMP, involved in maltosaccharide uptake and utilization, respectively (1, 2, 3). MalR belongs to the LacI-GalR family of transcriptional repressors (1) and binds specifically to two operators sequences (2) located in the intergenic region between the operons malXCD and malMP. Purified MalR protein was shown to bind more tightly to the malMP operator sequence (OM) than to the malXCD (OX), even though both operators differ only by two nucleotides. The binding of MalR to its DNA target is inactivated by the addition of maltose (2). MalR and PurR (another member of the LacI-GalR family) share an operator sequence and a HTH motif, involved in DNA binding, closely related (2), suggesting a common mechanism for DNA interaction. In that sense, a MalR molecular modelling by homology of PurR crystal structure (5) have been performed. The model show a good fit between both repressors specially in the DNA binding domain. 5 Several MalR mutants were isolated in S. Lacks’ laboratory (4). The repression and the allosteric capacity of these mutants were analysed and classified in two groups: i) constitutive mutants (Rc), in which the levels of amylomaltase (MalM) stay always very high and ii) Non inducible mutants (Rn), that are unable to grow in maltose and have the levels of MalM severely reduced. We have determined, in every mutant, the changes in the amino sequence and the position of these mutations on the MalR modelled structure. The DNA binding capacity of all these mutants also have been analysed. Among all the mutants, MalRn7 showed a most interesting behaviour. The mutation malRn7 is located outside from the DNA binding domain, but MalRn7 bound to OM and OX operator with higher affinity than wild type protein, increasing the repression activity “in vivo” over malXCD and malMP operon. A structural model to explain this superepressor activity will be discussed. References: 1. Puyet, A. , Ibañez, M., and Espinosa, M.(1993). Characterization of the Streptococcus pneumoniae maltosaccharide regulator MalR, a member of the LacIGalR family of repressors displaying distinctive genetic features. J.Mol.Biol. 268: 25402-25408. 2. Nieto, C., Espinosa, M., and Puyet, A. (1997). The maltose/maltodextrin Streptococcus pneumoniae. J. Biol. Chem.272: 30860-30865. regulon of 3. Nieto, C. Puyet, A. and Espinosa, M. (2001). MalR-mediated regulation of the Streptococcus pneumoniae malMP operon at promoter PM. J.Mol.Biol. 276: 1494614954. 4. Lacks,S. Genetic regulation of maltosaccharide utilization in Pneumococcus. (1968). Genetics 60: 685-706. 5. Schumacher, M.A., Choi, K. Y. , Zalkin, H. Brennan, R. G. (1994). Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by helices. Science 266: 763-770. Pneumococcal two-component systems as targets for the discovery of novel anti-bacterials and anti-infectives. Jerry Wells 6 Gene regulation by the PnpR/S two component system of Streptococcus pneumoniae. J. McCluskey, K, Overweg, J, Wells & T.J. Mitchell. Division of Infection & Immunity, University of Glasgow, Joseph Black Building, South Lab, University Avenue, GLASGOW, G12 8QQ. The ability of bacteria to sense and adapt to environmental stimuli is often mediated by two component signal transduction systems (TCSs). Global analysis of the genome of Streptococcus pneumoniae revealed 13 TCSs. In this study we examined pneumococcal TCS04 which consists of a response regulator and a histidine kinase encoded by pnpR and pnpS, respectively. Homologues of this system are present in other bacteria and have been shown to up-regulate the expression of many genes in response to low environmental phosphate levels. Included are the phosphate specific transporter genes pstS, C, A, B. Previous investigations have revealed that the pneumococcal pnpR/S and the pstSCAB genes are adjacent on the genome but unlike in other bacteria, no regulation of the transporter genes by the TCS has been demonstrated. This investigation examined the association of both these genetic loci in more detail. Microarray technology and proteomic analysis was used to compare pnpR/S mutants with the wild type. Analysis of the microarray and proteomic data will enable us to determine which pneumococcal genes may be regulated by this two component system. Genes of interest were analysed further using by RT-PCR and northern blotting. Purification and Polar Localization of the Pneumococcal LytB -NAcetylglucosaminidase: the Chain-Dispersing Murein Hydrolase. López, R., de las Rivas, B., García, J. L. and García, P. Centro de Investigaciones Biológicas, CSIC, Velázquez 144, 28006 Madrid, Spain. The DNA region encoding the mature form of a pneumococcal murein hydrolase (LytB) has been cloned and expressed in Escherichia coli. LytB was purified by affinity chromatography as a 74-kDa protein and its activity was assigned to be the first identified -N-acetylglucosaminidase of Streptococcus pneumoniae. LytB can only remove a maximum of 25% radioactivity from [methyl-3H]-labeled choline pneumococcal cell walls in in vitro assays. Inactivation of the lytB gene of the wild-type strain R6 (R6B mutant) led to the formation of long chains but did not affect either autolysis at the stationary phase of growth or development of genetic competence. Longer chains were formed when the mutation lytB was introduced into the M31 strain (M31B mutant), which harbors a complete deletion of lytA coding for the major autolysin. Purified LytB added to pneumococcal cultures of R6B or M31B was capable of dispersing, in a dosedepending manner, the long chains characteristic of these mutants into diplo cells or short chains, a morphology typical of R6 or M31 strains, respectively. In vitro acetylation of purified pneumococcal cell walls does not affect the activity of LytB whereas that of the 7 LytA amidase, the major murein hydrolase of pneumococcus, was completely abolished. The use of a translational fusion constructed between gfp and lytB supports the notion that LytB accumulates at the cell poles either of the wild-type R6 or the lytB mutants. This fusion protein was also able to unchain the lytB mutant. These observations propose the existence of specific LytB receptors that are positioned at the polar region on the pneumococcal surface. 5’ Nuclease of pneumococal DNA polymerase I. Active site and mechanism of action Mónica Amblar and Paloma López Centro de Investigaciones Biológicas, Madrid Spain The DNA polymerase I from Streptococcus pneumoniae (Spn PolI) is a bifunctional protein having two enzymatic activities: DNA polymerase and 5’ nuclease. These activities are located on different domains of the protein and, like in other type-I like DNA polymerases, both of them are involved in DNA replication and repair processes. Sequence comparison and enzymatic studies have shown that the eubacterial Pol I associated 5’ nucleases share significant sequence homology and several functional aspects with polymerase-independent 5’ nucleases from several bacteriophages and mammalian cells. The active site of these enzymes consists of a set of carboxylate residues that are highly conserved in all 5’ nucleases. This residues coordinate metal ligands that are essential for the nuclease activity. In order to establish the key residues of the Spn PolI exonuclease activity, a set of exonuclease mutants were constructed. The exonuclease and polymerase activities of the purified enzymes were analysed. The DNA binding capability and the exonuclease metal dependency, as well as, the catalytic rates for the exonuclease activity, were determined. The results obtained allowed us to identify some essential residues for the catalytic event and for the metal binding at the active site of the exonuclease domain. Moreover, a three-dimensional model of the Spn PolI exonuclease domain was built based on the available structural data from other related nucleases. Based on the experimental results together with the 3-D model, we proposed a possible active site of the Spn PolI exonuclease domain and a mechanism for the exonucleolytic reaction that is valid for all prokaryotic and eukaryotic 5’ nucleases. Molecular peculiarities of the lytA gene isolated from clinical pneumococcal strains that are bile-insoluble Garcia, J.L.,1 Obregon, V.,1 Garcia, P.,1 Garcia, E.,1 Fenoll, A.2 and Lopez, R.1 1 Centro de Investigaciones Biologicas, CSIC, Velazquez 144, Madrid, and 2Centro Nacional de Microbiología, Instituto de Salud Carlos III, Majadahonda, Madrid 8 The autolytic LytA amidase from 12 bile (deoxycholate)-insoluble streptococcal isolates (formerly classified as "atypical" Streptococcus pneumoniae) showing different antibiotic resistance patterns has been studied. These atypical strains, which autolyse at the end of the stationary phase of growth, contain highly divergent lytA alleles (pairwise evolutionary distances of about 20%) when compared to the lytA alleles of typical pneumococci. The atypical LytA amidases exhibit a peculiar deletion of two amino acids in the carboxy-terminal domain responsible for cell wall anchoring and have a reduced specific activity. These enzymes were inhibited by 1% deoxycholate but were activated by 1% Triton X-100, a detergent that could be used as an alternative diagnostic test for this kind of strains. Preparation of functional chimeric enzymes, PCR mutagenesis, and gene replacements demonstrated that the characteristic bile insolubility of these atypical strains was due to their peculiar carboxy-terminal domain, and that the 2 amino acids deletion was responsible for the inhibitory effect of deoxycholate. However, the deletion alone did not affect the specific activity of LytA. A detailed characterization of the genes encoding the 16S rRNA and SodA together with multilocus sequence typing indicated that the strains studied here are not a single clone and, although they cannot be strictly classified as typical pneumococci, they represent a quite diverse pool of organisms closely related to S. pneumoniae. The clinical importance of these findings is underlined by the role of lytA gene in shaping the course of pneumococcal diseases. This study can also contribute to solve diagnostic problems and to understand the evolution and pathogenic potential of species of the mitis group. A genomic and functional overview of surface active peptidyl-prolyl isomerases of Streptococcus pneumoniae Peter V. Adrian1, Alison Kerr2, Theo Hoogenboezem1, Ronald de Groot1, Tim J. Mitchell2, Peter W.M. Hermans1 Department of Pediatrics, Sophia Children’s Hospital, Erasmus University Rotterdam, The Netherlands, and 2 Division of Infection and Immunity, University of Glasgow, UK 1 Peptidyl-prolyl isomerases (PPIase) are ubiquitous house-keeping chaperones which catalyze the rate limiting cis-trans conformational changes at Xaa-Pro bonds during protein folding. Motive searches of the TIGR genome for all three known PPIase types (cyclophilins, FKBPs, and parvulin) revealed the presence of four enzymes containing putative PPIase domains. Two cyclophilins, of which one forms the C-terminal half of a putative cytoplasmic protein of unknown function, another is a lipoprotein and has been designated SlrA (Streptococcal lipoprotein rotamase A). In addition, a putative trigger factor homologue, and a parvulin like lipoprotein, previously identified as PpmA were identified. PPIase assays with an Xaa-Pro containing oligopeptide showed that recombinant SlrA exhibited high levels of PPIase activity. PPIase activity for recombinant PpmA was undetectable. The apparent lack of PPIase activity for PpmA is 9 not altogether unexpected since the low PPIase activity E. coli homologues SurA and PpiD are able to function as chaperones, without an active PPIase domain. While PpmA has been shown to play an important role in virulence, its function remains an enigma. In vitro, PpmA negative mutants are similar in cell and colony morphology, growth rate at different temperatures, ability to develop genetic competence, rate of autolysis, and susceptibility to a variety of chemical agents, antibiotics and surfactants. Failure to detect any differences may be due to a functional overlap with SlrA. The role of SlrA in virulence is currently being investigated. The membrane-associated F0F1 ATPase is essential for the viability of Streptococcus pneumoniae ADELA G. DE LA CAMPA* AND MARÍA JOSÉ FERRÁNDIZ Centro Nacional de Biología Fundamental, Instituto de Salud Carlos III, 28220 Majadahonda, Madrid, Spain. Despite the knowledge of pneumococcal genome sequences, little is known about their essential genes that could be targets of new antimicrobial compounds. The pneumococcal F0F1 ATPase, which is involved in intracellular pH regulation, shows a unique sensitivity to amino alcohol antimalarial drugs. Genetic studies aimed at eliminating expression of the atp operon encoding this enzyme by disruption of atpC, the first gene of the operon, were performed. An atpC::cat fusion in which atpC was interrupted by the chloramphenicol-resistance (Cmr) cassette (CRC: Ptet and Pcat promoters- cat genetranscriptional terminator) of plasmid pJS3, was constructed. Since the operon is transcribed from the single Patp promoter, chromosomal disruption of atpC implies that of the whole operon, given that transcription of the downstream atp genes, either from Patp, Ptet or Pcat would stop at the transcription terminator located in CRC. No Cmr transformants were obtained when this atpC::cat fusion was used in transformation experiments with the aim of substituting atpC into the R6 chromosome. Resistant transformants were obtained only when the recipient strain had a duplication of atpC. Three of these transformants showed a replacement of atpC by atpC::cat into the atpC copy located immediately upstream of the atp operon, in such a way that transcription of the operon from its own promoter was allowed. Given the requirement of the F0F1 ATPase for cell survival, it could be considered a target for the design of new antibacterial compounds, and invaluable as a model in the study of new antimalarial agents. The F0F1 H+-ATPase of Streptococcus pneumoniae is the target of mefloquine and new related compounds. ANTONIO JAVIER MARTÍN-GALIANO1*, 10 BEGOÑA GORGOJO1, CALVIN KUNIN2, AND ADELA G. DE LA CAMPA1 Centro Nacional de Biología Fundamental, Instituto de Salud Carlos III, 28220 Majadahonda, Madrid, Spain, 1 and Department of Internal Medicine, The Ohio State University, Columbus, Ohio. 2 The activity of mefloquine (Mef) and related compounds on previously characterized Streptococcus pneumoniae strains carrying defined amino acid substitutions in the c subunit of the F0F1 H+-ATPase was studied. In addition, a series of MefR strains were isolated and characterized. A good correlation was observed between inhibition of growth and inhibition of the membrane-associated F0F1 H+-ATPase activity. Mef was about 10fold more active than optochin and about 200-fold more active than quinine in inhibiting both growth and ATPase activities of the laboratory pneumococcal strain R6. Mutant strains showed different degrees of inhibition by the different compounds, depending on their specific mutations at the c subunit. The resistant strains studied here had point mutations that change amino acid residues either in the c or a subunits of the F0 complex. Changes in the c subunit were located in one of the two transmembrane -helices: residues M13, G14, G20, M23 and N24 of helix-1; residues M44, G47, V48, A49 and V57 of helix-2. Changes at the a subunit were also found in either of the transmembrane -helices 5 or 6: residue L186 of helix 5; residues W206, F209, and S214 of helix 6. These results suggest that the transmembrane helices of the c and a subunits interact and that the mutated residues are important for the structure of the F0 complex and proton translocation. The molecular basis of high frequency capsule phase variation on S. pneumoniae R.Waite, K. Struthers, C.G.Dowson: University of Warwick The molecular genetic basis of high frequency serotype 3, 8 and 37 capsule phase variation in Streptococcus pneumoniae was investigated. Pneumococci were grown in sorbarod biofilms at 34oC to mimic nasopharyngeal carriage. Different pneumococci generated apparently random tandem duplications of 11 to 239 bp segments of a single ORF within the capsule locus. These duplications alone were found to be responsible for high frequency capsule phase variation, where (phase off) acapsular variants possessed duplications within cap ORFs, and (phase on) capsulate revertants that possessed WT ORFs, indicating the precise excision of the duplication. Additionally, the frequency of phase reversion (off to on) was found to exhibit a linear relationship between (log) frequency of reversion and (log) length of duplication. This apparently random duplication giving rise to phase variation is in stark contrast to the ‘pre-programmed’ contingency genes in many Gram negative organisms that possess homopolymeric sequence repeats or motifs for site specific recombination. 11 Upper and lower respiratory tract infection by Streptococcus pneumoniae is affected by deficiency of pneumolysin and by differences in capsule type. Aras Kadioglu, Francesco Iannelli2, Gianni Pozzi2, Peter W. Andrew Department of Microbiology & Immunology, University of Leicester, England. 2 Laboratory of Molecular Microbiology and Biotechnology, Department of Molecular Biology, University of Siena, Siena, Italy. Streptococcus pneumoniae is an important respiratory pathogen of humans causing pneumonia, septicaemia, meningitis and otitis media. The pneumococcal toxin pneumolysin is heavily implicated in triggering inflammation and toxaemia that accompany these diseases. Pneumococci frequently colonise the upper respiratory tract where they are believed to act as a reservoir of infection of the lower respiratory tract and bacteraemia. We investigated how the pneumococcal toxin pneumolysin affected the capability of the pneumococcus to infect the upper and lower respiratory tract in the mouse. Methods: To study differences in colonisation and disease MF1 mice were intranasally challenged with wildtype Streptococcus pneumoniae serotypes 2 (strain D39) and 3 (strain A66), a serotype 2 pneumolysin-deficient mutant (PLN-A), and a serotype 2 pneumolysin gene re-inserted mutant (Pn+). In addition we also examined a pneumococcal chimeric mutant in which capsule we switched from 2 to 3 (FP50) to gain further insight into the role capsule had to play in nasopharyngeal infection. Colonisation and growth characteristics of these pneumococci in nasopharyngeal, tracheal and lung tissue were studied over 24 hr period. Results & Conclusions: PLN-A pneumococcal numbers in nasopharyngeal, tracheal and lung tissue were significantly reduced as compared to wildtype serotypes 2 and 3, FP50 and Pn+ (P< 0.05). Differences in pneumococcal capsule type were also seen to have significant effects on pneumococcal infection of nasopharyngeal, tracheal and lung tissue. However, it was the combination of capsule type and genetic background that was important and the influence of this combination varied with the site of infection. The combination of capsule type and genetic background also determined virulence. Thus the wildtype serotype 3 strain was virulent whereas the capsule switched mutant was avirulent. Importantly though, absence of pneumolysin was found to be associated with significantly lower pneumococcal numbers in both the upper and lower respiratory tract. Interaction of the surface displayed pneumococcal -enolase with human plasmin(ogen) is mediated via a novel binding motif as revealed by biochemical analysis 12 S. Bergmann, O.Diekmann, R. Frank, G.S. Chhatwal, S.Hammerschmidt* German Research Centre for Biotechnology (GBF), Microbial Pathogenicity, Braunschweig Binding of human plasminogen and its subsequent activation promotes penetration of Streptococcus pneumoniae through reconstituted basement membranes. Pneumococci bind human plasmin(ogen) via the glycolytic enzyme –enolase. Electron microscopic studies indicated the presence of Eno on the pneumococcal surface and its capacity to reassociate to the bacterial surface, although no signal sequence and no features required for anchoring were present. Several earlier binding studies suggested a critical role of the carboxyterminal lysines in plasmin(ogen) binding. However, analysis of plasmin(ogen) and kringle binding to wildtype Eno and carboxyterminal modified Eno proteins using the surface plasmon resonance technique (SPR) suggested a further binding domain for plasmin(ogen) in Eno. Therefore, plasminogen-binding activity of pneumococci incubated either with wildtype Eno or modified Eno proteins was determined. The results confirmed binding activity of both wildtype and modified Eno indicating the critical role of a second binding motif resulting in plasminogen acquisition under native conditions. Analysis of spot-synthesized peptides of the complete Eno sequence, consisting of 15 amino acids each, suggested the presence of an internal binding motif between amino acids 246 and 260. Further spot membrane analysis using synthetic peptides of decreasing length of the identified 15-amino-acid motif identified an octapeptide as the minimal binding motif pivotal for plasmin(ogen) binding. Inhibition assays using a synthetic peptide of the identified motif which is present in Eno with highest surface probability will confirm the functional activity of this internal plasmin(ogen)-binding domain. The acquisition of plasminogen via Eno and its subsequent activation can be used to facilitate the penetration of pneumococci through biological membranes. Allelic variation in the highly polymorphic locus pspC of Streptococcos pneumoniae Francesco Iannelli, Marco R. Oggioni, Gianni Pozzi* LA.M.M.B, Dipartimento di Biologia Molecolare, Università di Siena, 53100 Siena, Italy PspC, also called SpsA, CbpA, PbcA, and Hic is a surface protein of S. pneumoniae studied for its antigenic properties, its capability to bind secretory IgA, C3 and complement factor H, and its activity as an adhesin. In this work we characterized the pspC locus of 43 pneumococcal strains by DNA sequencing of PCR fragments. Using PCR primers designed on two unrelated ORFs, flanking the pspC locus, it was possible to amplify the pspC locus of each of the 43 strains of S. pneumoniae. In 37 out of 43 strains there was a single copy of the pspC gene, while two tandem copies of pspC were found in the other 6 strains. The sequence of the pspC locus was different in each of the 43 strains. Insertion sequences were found in the pspC locus of 11 out of 43 strains. Analysis of the deduced amino acid sequence of the PspC variants showed a common organization of the molecules: (i) a 37-amino acid leader peptide which is conserved in all proteins, (ii) a N13 terminal portion which is essentially alpha-helical, and is the result of assembly of 8 major sequence blocks, (iii) a proline-rich region, and (iv) a C-terminal anchor responsible for the cell surface attachment. By sequence comparison we identified 11 major groups of PspC proteins. Proteins within one group displayed only minor variations of the amino acid sequence. An unexpected finding was that PspC variants could differ in the anchor sequence. While 32 of the PspC proteins displayed the typical choline binding domain of pneumococcal surface proteins, 17 other PspC showed the LPXTG motif, which is typical of surface proteins of other Gram-positive bacteria. This major difference in the anchor region was also observed in the adjacent proline-rich regions which differed considerably in size and composition. The Role of a Zinc Metalloprotease in the Virulence of Streptococcus pneumoniae C.E.BLUE1, J-P. CLAVERYS2, T.J.MITCHELL1 1 Division of Infection and Immunity, Institute of Biomedical and Life Sciences, Joseph Black Building, University of Glasgow, Glasgow, Scotland, U.K., G12 8QQ 2 UMR5001 Centre National de le Recherche Scientifique-Universite Paul Sabatier, Labotatiore de Microbiologie et Genetique Moleculaire, 118, Route de Narbonne, 31062 Toulouse Cedex, France Zinc metalloproteases have been identified as virulence factors in several human pathogens, including the elastase of Pseudomonas aeruginosa and the major secretory protein of Legionella pneumophila. Several bacterial toxins are also zinc metalloproteases. There are also many other zinc metalloprotease enzymes that are implied to have some role in bacterial pathoenicity. Streptococcus pneumoniae is known to possess a zinc metalloprotease gene (zmpB) that has recently been mutated, in strain R6, using mariner mutagenesis and analysed in vitro (Berge et. al. 2001). Mutagenesis by the mariner technique resulted in a range of truncations within the zmpB gene. We have transferred two of these gene mutations into the capsulated, virulent parental strain of R6, D39, and analysed them in our murine model of pneumococcal pneumonia and septicaemia. Loss of the zinc metalloprotease gene resulted in significantly lower levels of bacteraemia and increased survival time following both intranasal and intravenous challenge, when compared to the isogenic parental strain. This identifies ZmpB as a novel pneumococcal virulence factor. The zmpB gene is located downstream of a pneumococcal two-component signal transduction system, and a link between this system and regulation of this gene is currently being investigated, together with potential mechanisms of action of the enzyme in vivo. 14 The Yin and Yang of microbial virulence and host immune response in determination of Streptococcus pneumoniae disease outcome. Yaffa Mizrachi Nebenzahl, Sarit Lifshitz, Sara Novick, Rachel Teitelbaum, Galina Feldman, Maxim Portnoi, Ron Dagan. Faculty of Health Sciences, Ben Gurion University of the Negev, Beer-Sheva, Israel Objectives: The study aimed to elucidate bacterial and host factors determining the outcome of S. pneumoniae (Pnc) infection. Methods: Inbred mice were inoculated intranasally with Pnc serotypes 3 or 14. Bacterial load, morbidity, mortality, pathological changes of the lungs, and cytokines mRNA levels (TNF -10, IL12 and INF Results: Serotype 3 induced sepsis and death within 3 days post-inoculation of both BALB/c (Th2) and C57BL/6 (Th1) mice. Serotype 14 induced development of diffuse pneumonia in C57BL/6 or localized lymphocyte infiltration of the lung in BALB/c. Lung injuries spontaneously resolved. Mice demonstrated background levels of TNF and TGF and denovo synthesis of IL-10 mRNA characterized sepsis induced by Pnc serotype 3. Inoculation with Pnc 14 was accompanied by significant (p<0.01) decrease of TNF, TGFIL -10 mRNA expression, starting from 3 h postinoculation. Conclusions: Pnc virulence determines the type of the disease developed, while the host immune response defines its severity. Lethal disease is characterized by expression of immunosuppressive and inflammatory cytokines, resulting in inability of the host to mount an adequate immune response. Down-regulation of TNFand TGF immunological and clinical course of disease. The crystallisation and structural determination of pneumococcal virulence factors Alan Riboldi Tunnicliffe University of Glasgow Streptococcus pneumoniae is a bacterial pathogen that affects children and adults throughout the world and is the leading cause of illness and death in infants, the elderly and immunocompromised patients. The bacteria is responsible for diseases such as pneumonia, bacteraemia and meningitis and causes millions of deaths each year. Antibiotic resistance is becoming an increasing problem with many strains of S. pneumoniae resistance to a wide range of antibiotics such as vancomycin and erythromycin. Due to such problems, new, novel targets for antibacterial agents are being sought. 15 Such a target is the two-component signal transduction system, 13 of which exist in S.pneumoniae. Two-component systems are comprised of two distinct protein components. The histidine protein kinase (HPK) is anchored to the cell membrane and includes an extracellular sensor domain, a transmembrane region, a nucleotide binding domain and a dimerisation domain which contains the histidine residue which is phosphorylated by ATP when a stimuli is received. The second component is the cytoplasmic response regulator (RR). Again, this protein is comprised of distinct domains: The receiver domain which accepts the phosphoryl group on an aspartic acid from the HPK and the DNA binding, output domain which controls gene expression. We have expressed and purified three response regulator proteins both entire and receiver domains (RR02, RR06 and RR10). RR02 is essential for bacterial survival as viable knockout mutants could not be obtained in animal models. RR06 has been shown to up-regulate the pneumococcal virulence factor CbpA which is involved in adherence to mucosal surfaces leading to prolonged and more serious pneumococcal infection. RR10 has been shown to be involved in vancomycin tolerance and has significant homology to VanR from Enterococcus faecalis. Sequence analysis of MM1, a temperate bacteriophage of the type 23F Spanish/USA multiresistant epidemic clone of Streptococcus pneumoniae García, P., Obregón, V., García, E., López, R. and García, J. L. Centro de Investigaciones Biológicas, CSIC, Velázquez 144, 28006 Madrid The presence of temperate phages in the chromosome of clinical isolates of pneumococcus has been well documented in the recent literature, although a role of phages in virulence of this important pathogen remains to be established. We have completely sequenced the genome of MM1, a temperate phage of a multiresistant pneumococcal clone. MM1 belongs to the Siphoviridae family and its DNA has been isolated as a DNA-protein complex. The phage genome is terminally redundant, circularly permuted and contains 40,248 bp that encode 53 ORFs organized in five functional modules. We have determined the attachment sites and the phage integrase required for site-specific recombination of the integration process. The lysis region is formed by an N-acetylmuramoyl-L-alanine amidase gene and two ORFs that possibly code for a holin and anti-holin and are involved in the liberation of the amidase to the cell wall. We have also analysed the repressor that controls the lysogenic cycle and, on the other hand, we have found a putative cytosine-methylase of GATC sequences, which appears to be the first case of a DNA-modifying enzyme encoded by a pneumococcal phage. 16 Functionally Consistent Sequence Blocks Contribute to Clone Variability in Streptococcus pneumoniae Marco R Oggioni and Gianni Pozzi LA.M.M.B., Dipartimento di Biologia Molecolare, Università di Siena After the first genomic sequences of S. pneumoniae were available it appeared that the genetic variability between strains was quite extensive. Whole blocks of sequences appeared to be clone specific. These blocks contained even up to 20 ORFs, which often appeared to be complete operons. All together the amount of clone specific sequences added to more than 10% of the total genome. Recently the availability of more genomic sequences (3 complete and 2 partial genomes) enabled a more precise analysis. Comparative genome analysis demonstrates now that the above recognised sequence blocks do not represent clone specific sequences, but should be more precisely regarded as sequence blocks of a common pneumococcal gene pool, which are present in some strains and not in others. These blocks, even if not mobile by themselves, behave like mobile elements (plasmids, phages, conjugative transposons) due to the natural competence for genetic transformation, which permits efficient integration of heterologous DNA by double crossing over. Data now would allow to define a common pneumococcal genome which harbours both regions of high allelic divergence (capsule, PspC, PspA, IgA protease and others) and regions which may be alternatively present or absent in single strains. Correlation of this variability to bacterium-host interaction can now be more precisely investigated. Whole-genome and functional organization of miniature transposable elements in Neisseriae Chiara Abrescia, Eliana De Gregorio and Pier Paolo Di Nocera. Dipartimento di Biologia e Patologia Cellulare e Molecolare Università degli Studi di Napoli Federico II, Via S. Pansini 5, 80131 Napoli The chromosome of pathogenic Neisseriae is peppered by members of an abundant family (2% of the genome) of small DNA sequences (70 to 160 bp) known as Correia or nemis (for neisseria miniature insertion sequences) elements. Nemis feature long terminal inverted repeats (TIRs), and are frequently found close to cellular genes. In vivo and in vitro data let us establish that nemis are cotranscribed with cellular genes and processed, at either one or both TIRs, by the RNase III. In silico analyses revealed that the number of nemis is comparable in the N. meningitidis Z2491 (A serogroup) and the MC58 (B serogroup) strains, but is sharply reduced in the N. gonorrhoeae strain F1090. Consequently, several genes conserved in the gonococcus and the meningococcus are flanked by nemis DNA in the meningococcus genome only. The hypothesis that nemis may contribute to some of the pathogenic traits of the meningococci is reinforced by PCR 17 analyses, indicating that at least 30 different genes are flanked by nemis in N. meningitidis strains (filled sites), but not in N. lactamica strains (empty sites). Penicillin resistance in Streptococcus pneumoniae: PBPs, cell wall alterations and biological price. A. Gilbey, A. Lloyd, T. Bugg, C.G.Dowson ; University of Warwick Since its detection in the late 1960s penicillin resistance in Streptococus pneumoniae has become increasingly prevalent worldwide. These highly penicillin resistant isolates are invariably cross- resistant to other ß-lactam antibiotics due to alterations in three key penicillin-binding-proteins (PBPs) 1A, 2X and 2B. However, resistance may come with a price. Previous work has shown that the cell walls of penicillin-resistant pneumococci differ radically from those of typical penicillin susceptible isolates. This study set out to further examine the molecular genetic basis of cell wall alterations in penicillin resistant pneumococci and to determine the biological price of penicillin resistance by the construction of isogenic transformants. A New Clonal International Penicillin Resistant Serotype of Streptococcus pneumoniae. MICHAEL R. JACOBS1*, MOSES L. JOLOBA1, ELIZABETH PALAVECINO1, SARALEE BAJAKSOUZIAN1, ANNE WINDAU1, PETER C. APPELBAUM2 1 Case Western Reserve University and University Hospitals of Cleveland, Cleveland, OH; 2Hershey Medical Center, Hershey, PA. Most penicillin resistant pneumococi belong to serogroups 6, 9, 14, 19 and 23, which belong to well-recognized international clones, and are included in the recently licensed 7-valent pneumococcal conjugate vaccine. We found three clonal penicillin resistant (but otherwise susceptible) type 29 strains from US otitis media patients in 1997-1998. We searched our surveillance databases for isolates with the same phenotype, and serotyped these isolates. Type 29 strains were further characterized by PFGE. Of over 8,000 pneumococci in our 1997-2000 databases, 150 had this susceptibility pattern. Fifty-one of these were found to be serotype 29, from patients in the USA (2 in 1997, 1 in 1998, 5 in 1999 and 25 in 2000), 9 from Mexico (2 in 1999, 7 in 2000), 7 from Spain in 1999, and 2 from Canada (1 each in 1999 and 2000). Penicillin MIC50/90 values were both 2 mcg/ml. PFGE of representative isolates from each country and year were found to be identical or closely related. Isolates from Spain were from Valencia, while those from the USA were from 17 states, with largest numbers from Ohio, Texas and Washington. Isolates were predominantly from sputum specimens in adults, with only 11 isolates being from 18 children. The increasing prevalence of this clone of a previously rare and previously penicillin susceptible serotype in three countries in North America and one in Europe suggests that this clone is spreading. However, the origin of this clone is unclear. Inclusion of this serotype in future vaccines may be warranted. Effects of Amino Acid Alterations in Penicillin-Binding Proteins 1a, 2b, and 2x on Penicillin-Binding Protein (PBP) Affinity of Penicillin, Ampicillin, Amoxicillin, Cefditoren, Cefuroxime, Cefprozil and Cefaclor in 18 clinical isolates of Streptococcus pneumoniae. Peter C. Appelbaum1, Kensuke Nagai1, Michael R. Jacobs2* 1 Hershey Med Ctr, Hershey PA; 2 Case Western Res Univ., Cleveland OH Amino acid alterations in or flanking conserved motifs making up the active binding sites of penicillin-binding proteins (PBP) 1a, 2b, and 2x of pneumococci were correlated with changes in affinity of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil and cefaclor for these PBPs. Four penicillin-susceptible (PSSP), 8 -intermediate (PISP) and 6 –resistant (PRSP) pneumococci were studied by DNA sequencing of penicillin binding sites of pbp1a, 2x, and 2b genes of strains, and by determining 50% inhibitory concentrations (IC50s) of the 7 agents for PBP1a, 2x, and 2b. Two PSSP strains had alterations in PBP2x (L546V) (1 strain) or PBP2b (T445A) (1 strain). All 8 PISP strains had at least two alterations - T338P or A, or H394Y in PBP2X, and T445A in BPB2b. All PRSP strains had the same changes seen in PISP strains, as well as T371A or S substitutions in PBP1a. The two most resistant PRSP strains had a second change in PBP2x (M339F) in conserved motif. Affinity of penicillin and ampicillin for all three PBPs was decreased for PRSP and most PISP strains. Affinity of amoxicillin for PBP1a and 2x was only decreased for PRSP. Cefaclor and cefprozil showed decreased affinity of PRSP but not PISP for all three PBPs. Cefuroxime showed decreased affinity of PISP and PRSP for PBP1a and 2x, but no change for PBP2b. Cefditoren showed no difference in PBP affinity based on penicillin or cefditoren MICs, indicating a different PBP target for this agent. Overall, MICs and PBP affinities of the strains correlated with the changes found in the PBP active binding sites. Low-level resistance to rifampin in Streptococcus pneumoniae, molecular base and potential role for resistance development Kathrin Mühlemann, Silvia Utz, Patricia Stutzmann Meier Institute for Infectious Diseases, University of Bern, Bern, Switzerland 19 Background: In S. pneumoniae rifampicin resistance due to point mutations in the cluster I and III region of rpoB have been described. We report a mutation in the cluster II region mediating low-level resistance to rifampicin. Methods: Spontaneous, rifampicin-resistant mutants were generated in vitro by selection of susceptible, clinical isolates on blood agar plates containing rifampicin at concentrations of 0.5, 1, 4, 10 and 50 mg/l. A 2008bp fragment of rpoB was PCR amplified, sequenced and analyzed for point mutations. Results: A previously, non-published mutation in cluster II (T to A at position 1716 of E.coli rpoB gene) was observed in low-level resistant mutants from four different strains. Spontaneous high-level rifampicin resistant mutants had mutations in cluster I only. The link between low-level resistance and the mutation in cluster II was confirmed by transformation of a rifampicin sensitive strain with the PCR fragment harboring the point mutation in cluster II. Further selection of low-level resistant mutants on agar plates with increasing concentrations of rifampicin showed the acquisition of additional mutations in cluster I, although two mutants showed exclusively an additional C to A mutation at position 1717 in cluster II. Interestingly the mutation rates of the rifampicin mutants originating from low–level resistant S. pneumoniae were 10-fold higher than the mutation rates observed in rifampicin sensitive S. pneumoniae. Conclusions: Low-level resistance to rifampicin in S. pneumoniae is linked to a point mutation in cluster II and may be the first step towards acquisition of high-level resistance to this drug. Streptococcus pneumoniae macrolide resistance in Italy: characterization of the elements carrying the efflux genes mef(A) and mef(E) M. Del Grosso,1 F. Iannelli,3 M. Santagati,4 N. Petrosillo,2 S. Stefani,4 G. Pozzi,3 and A. Pantosti1 Laboratory of Bacteriology and Medical Mycology, Istituto Superiore di Sanità,1 Istituto Nazionale per le Malattie Infettive Lazzaro Spallanzani,2 Rome, LA.M.M.B., Dipartimento di Biologia Molecolare, Università di Siena, Siena,3 and Dipartimento di Scienze Microbiologiche, Università di Catania, Catania,4 Italy. The mef gene is a common determinant of macrolide resistance in Streptococcus pneumoniae encoding for a drug efflux pump. Susceptibility to macrolides was evaluated in a large series of erythromycin-resistant isolates from invasive diseases and healthy carriers. Of the 187 erythromycin-resistant strains, 40 (21%) showed an M phenotype and carried mef. Both mef(A) and mef(E) were found: 33 carried mef(A) and 7 mef(E). The characteristics of the strains carrying the mef genes and the properties of the mefcontaining elements were studied. All isolates carrying mef(A) belonged to serotype 14 and were susceptible to the antibiotics tested, except erythromycin. 17 mef(A)-positive strains were examined by PFGE. All shared very similar profiles suggesting they belong to the same clone. The allelic profile of one isolate, obtained by MLST (ST 9) corresponds to that of England14-9, a major antimicrobial-resistant clone. The mef(E) 20 strains belonged to six different serotypes, the majority were resistant to other antibiotics besides erythromycin, including penicillin. Three mef(E) strains examined by PFGE did not appear to be clonally related. The sequence of a fragment of the mef-containing element, encompassing mef and the msr(A) homolog, was respectively identical in the 3 mef(E)-positive strains and in 3 mef(A)-positive strains examined, although there were differences at 168 positions between the 2 groups. In all mef(A)-positive strains, the mef element was inserted in celB, that led to impairment of the competence of the strains. In line with insertion of the mef(E) element at a different site, the competence ability of the mef(E)-positive strains was maintained. Transfer of erythromycin resistance by conjugation was obtained from 2 out of 3 mef(A) strains but from none of 3 mef(E) strains. Due to the important different characteristics of the strains carrying mef(A) or mef(E), we suggest to maintain the distinction between the two genes. Vaccination and host response to Streptococcus pneumoniae in animal models of infection Tim Mitchell Division of Infection and Immunity, Institute of Biomedical and Life Sciences Joseph Black Building, University of Glasgow, Glasgow G12-8QQ, Scotland Discovery of protein vaccine antigen candidates against Group B streptococci using LEEP, a new technology for the rapid identification of genes encoding exported proteins in Gram-positive pathogens. S. B. Hanniffy1, R. Seepersaud2, P. J. Mayne1, P. Sizer3, R. Le Page2, J. Wells1; 1 Institute of Food Research, Infection and immunity Group, Norwich Research Park, Norwich, NR4 7UA, UNITED KINGDOM, 2Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge, CB2 1QP, UNITED KINGDOM, 3Provalis U.K. Ltd, Deeside Industrial Park, Deeside, Flintshire, UNITED KINGDOM. Streptococcus agalactiae (Group B Streptococcus; GBS) is the leading cause of neonatal bacterial infection in the developed world. While improved antimicrobial prophylaxis can reduce the incidence of early-onset GBS disease, it is unlikely to prevent late-onset infections, prematurity and stillbirths due to GBS. Our work is focused on vaccine antigen discovery and development for GBS. Surface-exposed GBS proteins which induce antibodies able to protect against infection by different capsule antigen serotypes would represent strong candidates for use in an GBS vaccine. In order to determine whether such proteins exist we have developed and used a new method for rapidly screening the genomes of Gram-positive pathogens for genes encoding exported proteins. This method, termed "LEEP" (= lactococcal expression of exported proteins) exploits the functionality of recombinant secretion leader capture vectors in Lactococcus lactis. The LEEP screening vectors incorporate a 21 lactococcal promoter and use staphylococcal nuclease as a secretion reporter: the export of this protein can be easily be detected on an indicator medium. Appropriately sized DNA fragments derived from GBS were inserted into LEEP vectors in 3 different reading frames, and the resulting plasmids were screened for their capacity to direct nuclease secretion in transformed L. lactis. Approximately 200 partial and fulllength gene sequences encoding novel putative GBS surface proteins were quickly recovered and identified. PCR methodology was used to determine the extent of allelic variation of recovered sequences amongst a panel of clinical isolates representing all 9 GBS serotypes. DNA vaccination-based protection data and bioinformatic analysis were used to screen and prioritise genes whose products might represent candidate immunogens. The leading candidates have now been tested as proteins in an adult mouse infection model and a significant number of the antigens possess protective activity; some of which display similarity to previously identified protective antigens from Streptococcus pneumoniae. Our results validate the use of LEEP for the identification of surface protein antigens likely to elicit protective immune responses which can be adapted for use with other Gram-positive pathogenic bacteria including S. pneumoniae The interplay between proinflammatory and inhibitory cytokine responses in determination of Streptococcus pneumoniae infection outcome Y. Mizrachi Nebenzahl E. Ling, N. Troyanovsky, G. Feldman, R. Dagan. Pediatric Infectious Disease Unit, Soroka Medical Center, Faculty of Health Sciences, Ben Gurion University of the Negev, Beer-Sheva, Israel Objectives: To analyze host factors active at the early stages of S. pneumoniae (Pnc) infection of the young animals. Methods: Models of Pnc infection of young animals were established. 3 weeks old C57BL/6 (Th1 type) and BALB/c (Th2 type) inbred mice were inoculated intranasally with Pnc serotype 6B or 14. Bacterial load and cytokine mRNA levels representing innate and specific immune response (TNF, TGF IL-10, IL-12) were analyzed. Results: Pnc serotype 6B colonized the nasopharynx, whereas serotype 14, along with colonization of upper respiratory tract, penetrated into the lungs. Infection with both serotypes was characterized by significant increase of TNF (p<0.05) and de-novo expression of IL12 and TGF starting 3 h post-inoculation. It should be noted, however, that serotype 14 induced expression of IL-10, whereas it was negligible upon Pnc 6B infection. Conclusions: Pnc infection of young animals is characterized by de-novo expression of proinflammatory cytokines, whereas expression of immunosuppressory cytokine IL10, seen in invasive infection, might stipulate the spread of Pnc into the young host. 22 PpmA of Streptococcus pneumoniae immunization experiments protects mice in passive Peter V. Adrian1, Alison Kerr2, Ronald de Groot1, Tim J. Mitchell2, Peter W.M. Hermans1 Department of Pediatrics, Sophia Children’s Hospital, Erasmus University Rotterdam, The Netherlands, and 2 Division of Infection and Immunity, University of Glasgow, UK 1 Introduction. We recently characterized the PrtM-like protein A (PpmA) of Streptococcus pneumoniae, a liproprotein which renders interesting properties with respect to its potential use in future conjugate vaccines. (i) Immuno-electron microscopy with rabbit antibodies raised against PpmA has demonstrated that this protein was surface-associated. (ii) The opsonophagocytic activity of anti-PpmA rabbit antibodies is high and species-specific. (iii) DNA sequence analysis of the ppmA genes of various genetically distinct pneumococcal strains has demonstrated limited variation. In this study, we investigated the immune-protective potentials of antibodies raised against recombinant PpmA in passive immunization experiments in mice. Methods. MF-1 mice (12 per group) were vaccinated by intraperitoneal injection of 200µl of compliment inactivated rabbit serum, 24h and 1h prior to infection. Control groups were vaccinated with PBS, pre-immune serum, and serotype-2 capsular antibodies. Mice were infected intranasally with 106 CFU of pneumococcal strain D39 (serotype 2) in 50l. Mice were monitored frequently during the following 14 days for signs of infection. On reaching the pre-determined endpoint (moribund), mice were sacrificed and the survival times were noted. Results. Mice injected with either anti-PpmA or anti-capsular serum and intranasally challenged with a lethal dose of pneumococci had significantly longer survival times (p<0.05) than those injected with pre-immune serum or PBS. Conclusion. The passive immunization data indicate that PpmA of S. pneumoniae has promising immune-protective potentials with respect to its use in future conjugate vaccines. These findings justify current active vaccination studies with this component. Adhesins as candidate protein vaccines for Streptococcus pneumoniae Y. Mizrachi Nebenzahl1, E. Ling1, G. Feldman1, S. Lifshitz1, M. Portnoi1, K. Overweg2, J. Wells2, R. Dagan1. 1 Faculty of Health Sciences, Ben Gurion University of the Negev, Beer-Sheva, Israel; 2 Institute of Food Research, Norwich, UK Pnc adhesion to host cell membrane is of critical importance for colonization and infection of the host. We hypothesized that Pnc lectin proteins elicit an immune response that will prevent Pnc adhesion and thus interfere with infection. Pnc cell wall (CW) proteins were separated into lectin (L) and non-lectin (NL) fractions by fetuin affinity chromatography. C57BL/6 and BALB/c mice were vaccinated with Pnc total CW, L and 23 NL protein preparations mixed with Freund’s adjuvant. Animals were challenged intranasally with 108 cfu or intraperitoneally with 107 cfu of Pnc serotype 3. 100% of the control sham vaccinated mice died within 96 hours post-inoculation. Vaccination with total CW protein preparation resulted in survival rates of 66 and 100% following intraperitoneal and intranasal challenges, respectively. Vaccination with CW-NL and CW-L resulted in survival rates of 55-66% and 33-50% following intranasal challenge, respectively; and in respective survival rates of 66-75% and 25-33%, following intraperitoneal challenge. 2D gel electrophoresis and MALDI-TOF allowed identification of 32 cell wall proteins present in these mixtures. The results indicate the ability of CWL proteins to partially interfere with Pnc infection. Non-lectin proteins may be important particularly against invasive disease. An efficient vaccine may need to combine immunization with adhesins and proteins expressed by bacteria in sepsis. POSTERS Surface plasmon resonance (SPR) analysis indicated the presence of a novel binding motif in the pneumococcal adhesin Eno for plasmin(ogen) S. Bergmann, O. Diekmann, R. Frank, G.S. Chhatwal, S. Hammerschmidt German Research Centre for Biotechnology (GBF), Microbial Pathogenicity, Braunschweig The glycolytic enzyme α-enolase has been identified as surface exposed pneumococcal adhesin for plasmin(ogen). The Eno designated protein is secreted and bound to the cell wall surface despite motifs such as the signal peptide and membran anchor are not present. Moreover, free Eno is able to reassociate to the pneumococcal cell surface thus enhancing the acquisition of plasminogen. The carboxyterminal lysines are known to be important mediators of the plasmin(ogen) interaction via binding to the lysine-binding sites of the kringle motifs. To analyse the Eno-plasmin(ogen) interaction Eno proteins with substituted or deleted carboxyterminal lysine residues immobilized on CM5 sensor chips were used to determine the reaction kinetics of plasmin(ogen) and kringle 1-3 binding to wildtype and modified Eno by surface plasmon resonance technique (SPR). The sensogram data of BIAcore analysis revealed no significant differences in the reaction kinetics between modified Eno proteins and wildtype Eno. Binding experiments with Eno proteins reassociated to pneumococci and radiolabeld plasminogen confirmed the binding activity of both wildtype and modified Eno proteins. To elucidate the binding motif spot synthesized peptides of Eno, each of 15 amino acids, were analysed for plaminogen binding. This approach identified a novel binding motif in Eno for plasmin(ogen) binding which is most probably surface exposed and thus should be accessible for the Eno-plasmin(ogen) interaction. 24 To confirm the critical role amino acid substitutions were introduced in the internal binding motif and the binding of plasmin(ogen) to mutated peptides and Eno proteins analysed. The results of these studies strongly indicate a pivotal role of the identified internal binding motif for the Eno-plasmin(ogen) interaction. Genomic Comparisons of Streptococcus spp. by microarrays. Reinhold Brückner1, Beate Weber1, Nadège Balmelle2, Christophe Gardès2, Wolfgang Keck2, Antoine de Saizieu2, and Regine Hakenbeck1. 1 Universität Kaiserslautern, Abteilung Mikrobiologie, Paul-Ehrlich-Str. 23, D-67663 Kaiserslautern, 2 Preclinical Pharma Research, F. Hoffmann-La Roche AG, CH-4070 Switzerland Streptococcus pneumoniae remains a major causative agent of serious human diseases. The worldwide increase of antibiotic resistant strains revealed the importance of horizontal gene transfer in this pathogen, a scenario that results in the modulation of the species-specific gene pool. We investigated genomic variation in 20 S. pneumoniae isolates representing major antibiotic-resistant clones and 10 different capsular serotypes.Variation was scored as decreased hybridization signals visualized on a highdensity oligonucleotide array representing 1,968 genes of the type 4 reference strain KNR.7/87 (TIGR4). Up to 10% of the genes appeared altered between individual isolates and the reference strain; variability within clones was below 2.1%. Ten gene clusters covering 160 kb account for half of the variable genes. Most of them are associated with transposases and are assumed to be part of a flexible gene pool within the bacterial population; other variable loci include mosaic genes encoding antibiotic resistance determinants and gene clusters related to bacteriocin production. Genomic comparison between S. pneumoniae and commensal Streptococcus mitis and Streptococcus oralis strains indicates distinct antigenic profiles and suggests a smooth transition between these species, supporting the validity of the microarray system as an epidemiological and diagnostic tool. The critical role of the hexapeptide binding motif of the pneumococal SpsA/CbpA protein in the interaction with the secretory component as revealed by surface plasmon resonance (SPR) Christine Elm1*, Manfred Rohde1, Jean-Pierre Vaerman2, G. Singh Chhatwal1, und Sven Hammerschmidt1 1 German Research Centre for Biotechnology (GBF), Microbial Pathogenicity, Braunschweig 2 Université Catholique de Louvain and Institute of Cell Pathology, Brussels 25 The poly-immunglobulin receptor (pIgR) of mucosal epthelial cells mediates the transport of pIgA across polarized epithelial cells, resulting in release of secretory component (SC), either free or bound covalently to IgA. A hexapeptide motif of pneumococcal surface protein SpsA, also designated CbpA (PspC), has been shown to interact in a human specific manner with the SC/pIgR. In vitro assays indicated that this interaction facilitates adherence in pIgR expressing epithelial cells. The interaction and species-specificity of the SpsA-SC/IgR interaction was analysed by surface plasmon resonance (SPR) on the BIAcore optical biosensor (Pharmacia Biosensor 2000). Native SpsA protein and recombinant SpsA-derivatives representing functional domains of SpsA as well as SpsA201 with a single amino acid substitution in the binding motif were used as ligands and immobilized on CM5 sensor chips. SC and SIgA from human, bovine, canine, guinea pig, hamster, mouse, rabbit and rat were used as analytes to verify the species-specificity. The data of the BIAcore analyses confirmed clearly the human species-specificity and suggested that the in vivo effects shown for colonization and invasion of epithelial cells in animal models are most probably not due to the SpsA-pIgR interaction. Moreover, the sensograms of the SpsA201 showed no binding of SC and SIgA confirming the critical role of the identified hexapeptide for the SpsA-human SC interaction. Fitting the sensogram data indicated a simple 1:1 binding of human SC and SIgA to covalently bound SpsA and SpsA derivatives. The global fit modeling of the simple 1:1 Langmuir kinetic revealed similar association and dissociation constants for SC and SIgA irrespective of the molecular size of the ligand SpsA and the number of binding motifs in SpsA. However, these results might not reflect the in vivo situation since latex beads coated with different SpsA-derivatives exhibited different binding activities to pIgR expressing cells. Vancomcin-tolerance in Streptococcus pneumoniae: absence of drug tolerance in new vncS deletion mutants. B. DESAI,1 W. COLEMAN1, L. L. GRINIUS2, and *D. A. MORRISON1. 1 University of Illinois at Chicago, Chicago, IL 60607, 2 Procter and Gamble Pharmaceuticals, Mason, Ohio 45040. Vancomycin tolerant strains form a new class of drug resistant Streptococcus pneumoniae. Many of these strains carry mutations that map to the locus of a twocomponent regulatory system with genes named vncR and vncS. It was reported that SPSJ01, a strain created by insertional disruption of vncS, is tolerant to vancomycin as well as to beta-lactams, cephalosporins, aminoglycosides, and quinolones (Novak, et al., Nature 399:590-93, 1999). The hypothesis was proposed that the tolerant phenotype is caused by failure of VncS to dephosphorylate VncR, leading to a failure to relieve repression of autolysin function. To explore this hypothesis, all but 61 codons of vncS were replaced by a Kan cassette, creating strain CP1292. To detect possibly subtle levels of tolerance, the response of CP1292 was determined at 0.25, 0.35, 0.5, 0.7, 1.0, 2.0, 5.0, 26 and 20 µg vancomycin per ml in rich media and in a chemically defined medium. The MIC was between 0.5 and 1 micrograms per ml, and the response of CP1292 was indistinguishable from that of its wild type parent, CP1250. Thus, it appears that the loss of VncS function per se does not necessarily create a vancomycin-tolerant strain. We infer that the tolerance reported for the pJDC9 insertion in vncS in strain SPSJ01 may depend on a specific recombinant form of VncS, or on some other special feature of that strain. Genetic plasticitiy of Streptococcus pneumoniae: formation of chromosomal rearrangements and a large inversion by transformation Anne-Marie GASC, Bernard MARTIN and Jean-Pierre CLAVERYS Laboratoire de Microbiologie et Génétique Moléculaire, UMR 5100 CNRS-Université Paul Sabatier, 118 route de Narbonne, 31062 Toulouse Cedex, France. The gene cluster (cap) involved in capsular formation in Streptococcus pneumoniae has been characterized at a molecular level for several serotypes. In all cases, except serotype 37, the cap cluster is placed between the dexB and aliA genes (reviewed García et al., 2000, Res Microbiol 151: 429-435). A single gene (tts) required for synthesis of type 37 homopolysaccharide was shown to be located outside the cap locus (Llull et al., 1999, J Exp Med 190: 241-251). The gene arrangement reported in the type 37 isolate was gpmAtts-IS1167-pyrDA, suggesting the existence of a genome reorganization because in laboratory strains such as R6, gpmA(-abcT) and (metK-)pyrDA are separated by ~528 kb (Hoskins et al., 2001, J Bacteriol 183: 5709-5717). [We adopted the following nomenclature for the various chromosomal regions: A-B and Z-Y correspond respectively to the metK-pyrDA and gpmA-abcT gene arrangements. According to this nomenclature, the gene arrangement in type 37 strains is Z-tts-IS1167-B.] With the aim of investigating new facets of the transformation-dependent plasticity of S. pneumoniae, we first characterized the gene arrangement of the putative complementary region in type 37, as metK-IS1167-abcT (i.e. A-IS1167-Y according to the above nomenclature). This observation allowed us to propose models to account for the formation of two previously described recombinants (Llull et al., ibid.), i.e. a type 37like recombinant and a recombinant harboring the gpmA-tts-IS1167-abcT (i.e. Z-//-Y) arrangement. We then used as donor in transformation chromosomal DNA from a type 37-like strain carrying a disruption of tts by the ermC gene which confers resistance to erythromycin (Ery). EryR transformants were selected and analyzed by PCR and Southern hybridizations. Results will be presented and discussed. Genetic Variability and Specificity of the Competence Two-Component System in Streptoccoccus pneumoniae 27 FRANCESCO IANNELLI, DONALD A. MORRISON, AND GIANNI POZZI. 1 LA.M.M.B., Dipartimento di Biologia Molecolare, Università di Siena, Siena, Laboratory for Molecular Biology, Department of Biological Sciences, University of Illinois at Chicago, Chicago Illinois 60607 2 Competence for genetic transformation in Streptococcus pneumoniae is regulated by a quorum-sensing mechanism involving the pheromone CSP (Competence Stimulating Peptide) and a two-component system, ComD/ComE. CSP is encoded by comC and is processed and exported by ComAB. It is assumed that the trans-membrane histidine kinase ComD in the presence of CSP activates the cognate response regulator ComE which induces the late competence genes. In this work we determined the sequence of the comD and comE genes in the pneumococcal strain A66, and we found a new allelic variant of comD, denominated comD2. The genetic diversity between comD and comD2 is restricted to the region encoding the sensor domain of the protein. The comD sensor domain coding sequence was also characterized in twelve additional pneumococcal strains, and we observed further variability. Synthetic pheromones were used to induce competence in isogenic comC-negative mutants containing three different ComD receptors. Regulation of comABCDE and the early control of competence for genetic transformation in Streptococcus pneumoniae Marc Prudhomme, Benoît Grossiord, Bernard Martin, and Jean-Pierre Claverys Laboratoire de Microbiologie et Génétique Moléculaire, UMR 5100 CNRS-Université Paul Sabatier, 118 route de Narbonne, 31062 Toulouse Cedex, France. It is now well established that the development of competence of Streptococcus pneumoniae is under the control of a two-component regulatory system (TCS), ComDE, and the comC encoded competence stimulating peptide (CSP) (Pestova et al., 1996, Mol Microbiol 21, 853-864; Håvarstein et al., 1995, PNAS 92, 11140-11144).The CSP is exported by the ComAB transporter. When sensing CSP, ComD activates ComE which controls expression of the competence regulon both directly and indirectly, through the alternative sigma factor, ComX (Lee and Morrison, 1999, J Bacteriol 181, 5004-5016). To address the question of whether CSP accumulates passively in the growth medium or whether its production can be temporarily increased in response to changes in environmental conditions, transposon insertion mutants upregulating comCDE (cup mutants) were isolated (Martin et al., 2000, Mol Microbiol 38, 867-878). The analysis of three classes of cup indicated i) that CSP export through ComAB is normally limiting, leading to the conclusion that comAB constitutes a potential target for regulation. ii) that a previously identified TCS system, CiaRH, controls directly or indirectly the expression of 28 comABCDE; iii) and revealed that ClpP, an ubiquitous stress response protease, acts negatively on competence development (Chastanet et al., 2001, J Bacteriol 183, 72957307). Other cup mutants indicating that modifications of purine metabolism, alterations of peptidoglycan synthesis and changes in membrane lipid composition affect competence development will be described. Collectively, these observations suggest that competence development, is adjusted by a complex network in response to changing environmental conditions. Identification of Pneumococcal Vaccine Antigens Using a Gram-positive Secretion Reporter Screen in Lactococcus lactis. D. B. Badcock1, P. Hansbro2, S. Hanniffy3, V. Clarke1, K. Schofield1, K. Robinson4, P. Sizer5, R. Le Page1, J. Wells3; 1 Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge, CB2 1QP, UNITED KINGDOM, 2Centre for Biomolecular Vaccine Technology, University of Newcastle, NSW2300, AUSTRALIA, 3Institute of Food Research, Infection and immunity Group, Norwich Research Park, Norwich, NR4 7UA, UNITED KINGDOM, 4Queens Medical Center, Nottingham, NG7 2UH, UNITED KINGDOM, 5 Provalis U.K. Ltd, Deeside Industrial Park, Deeside, Flintshire, CH5 2NT, UNITED KINGDOM. Most previously identified virulence determinants of pathogenic bacteria have been found to be exported or surface associated. Such proteins are good candidates as vaccine antigens or as targets for anti-infectives and anti-bacterial agents due to their role in mediating crucial interactions between the bacteria and their environment. In this study we have used a genetic screen termed LEEP (= lactococcal expression of exported proteins) to identify exported proteins from S. pneumoniae with the aim of identifying novel vaccine antigens. Our approach is based on a modification of an earlier method used for isolation of signal peptide sequences in L.lactis and identifies translationally coupled proteins that direct secretion of a signal peptide-deficient nuclease reporter gene. The LEEP vectors (constructed in all three reading frames with respect to the nuclease reporter gene) are based on the pTREP vector designed for use in L.lactis and contain a constitutive lactococcal promoter to ensure expression of translational fusion proteins. Partially digested pneumoccocal DNA was ligated into the LEEP vectors and used to transform L.lactis. Plate assays for extra-cellular nuclease activity were used to identify 90 nuclease-secretion positive clones from which pneumoccocal DNA was sequenced. Computer analysis of the LEEP genes using the TIGR pneumoccocal database and bioinformatics tools revealed that the majority of the LEEP genes contained motifs predicted to be involved in protein export or membrane translocation. Further studies are now underway to investigate the vaccine potential of selected protein antigens in a respiratory tract infection model. Our results indicate that LEEP system is a powerful functional assay for identifying candidate surface immunogens from Gram-positive bacteria. 29 Details of proteins identified using the functional LEEP assay will be presented and compared with the results from a bioinformatics analysis of the genome sequence. The implications for vaccine development will be discussed. The Effects of Pneumococcal Virulence Factors on Brain Ependymal Cilia. space18033 Hirst RA*+, Rutman A*, Andrew PW+, O'Callaghan C*. Departments of Child Health* and Microbiology and Immunology+, University of Leicester, Leicester LE2 7LX Densely ciliated ependymal cells line the ventricular surface and aqueducts of the brain, forming a barrier between cerebrospinal fluid, which is infected in meningitis, and neuronal tissue. We have established an ex-vivo model of ependymal ciliary beat frequency (CBF) to evaluate the effects of the pneumococcus and its virulence factors. Our data shows that that a number of pneumococcal virulence factors can inhibit ependymal CBF. Beating cilia were recorded by high-speed video camera at 37oC. Slowmotion playback allowed beat frequency and pattern to be determined. Rat ependymal cells were exposed (at times up to 120 minutes) to D39 pneumococci, purified pneumolysin, and hydrogen peroxide. The data showed that CBF was inhibited by pneumococci and their virulence factors in a rank order potency: Purified pneumolysin (150HU)>Wild type D39 (108cfu/ml)>Bacterial levels of hydrogen peroxide (100M). In addition, analysis of the ependyma by scanning electron microscopy after bacteria or virulence factor addition, showed morphological anomalies when compared to control tissue (data to be presented). In conclusion, infection of the cerebrospinal fluid with pneumococci gives rise to the classic symptoms of meningitis including raised intracranial pressure and changes in cerebral blood flow. Our data shows that as the pneumococci are circulated within the CSF they are likely to impair brain ependymal function. Moreover, virulence factor release after antibiotic lysis is likely to cause further damage to the brain epithelium. Identification and analysis of proteins expressed by Streptococcus pneumoniae cultured in human pooled serum. K. Overweg, M. Liebregts, F. Mulholland, J.W. Wells Institute of Food Research, Norwich, U.K. Streptococcus pneumoniae is an important human pathogen which causes meningitis, otitis media, sepsis, and pneumonia. The precise mechanisms by which the pneumococcus survives in the human host are not fully understood. Generally, it is believed that identification and characterisation of in vivo expressed genes will provide insight into the mechanisms that underlie bacterial pathogenesis. Furthermore, in vivo 30 produced factors are likely to be necessary for bacterial adaptation, growth and survival in the host. We have grown S. pneumoniae in human pooled serum (HPS) as a model of pneumococcal sepsis in order to identify the bacterial factors that might be expressed in invasive disease. To identify proteins that are expressed by S. pneumoniae growing in HPS, total cellular extracts of two different pneumococcal strains grown in HPS and a rich broth (Todd Hewitt Yeast Extract broth; THY-broth) were analysed by twodimensional gel electrophoresis. The analysis of about 300 protein spots under both growth conditions revealed 95 differences in protein expression of which 69 were increased in amounts and 26 decreased in amounts in the HPS grown bacteria. All 95 protein spots were excised, digested with trypsin and further analysed by peptide mass fingerprinting using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-tof). Pneumococcal DNA microarrays are being constructed and will be used to compare gene expression profiles of these cultures with that of the proteomics data. The role of the differentially expressed proteins in vivo will be discussed. Janus, an rpsL Cassette for Gene Replacement through Negative Selection in Streptococcus pneumoniae. C. K. Sung1, H. Li1, J. P. Claverys2, and D. A. Morrison*.1 1Laboratory for Molecular Biology, University of Illinois at Chicago, Chicago, IL, USA ; 2 LMGM, UMR5100 CNRS-Université Paul Sabatier, Toulouse, France Streptococcus pneumoniae, a widespread human pathogen associated with high rates of disease and mortality, is being used increasingly as a genetically tractable model pathogen for application of genomics to searches for new drugs and drug targets. Natural genetic transformation offers a direct route by which synthetic gene constructs can be placed into its chromosome. To date, these gene disruptions have commonly been made by inserting a drug resistance gene that provides direct selection of rare recombinants. While powerful, this method does have drawbacks. As design of strains with multiple mutations becomes more sophisticated, for example, an accumulation of drug markers in the mutated strains could become cumbersome, and possibly compromise interpretations of experimental results. Also, many important categories of mutation, such as missense substitutions and in-frame deletions usually confer no selectable phenotype. Thus, the lack of a general negative-selection marker has hampered the use of constructs that do not confer a selectable phenotype. To create such a marker, a 1.3-kb cassette was constructed comprising a kanamycin (Kn) resistance marker (kan) and a counterselectable rpsL+ marker. The cassette conferred dominant Sm sensitivity in a SmR background. The cassette was used in a two-step transformation procedure to place DNA of arbitrary sequence at a chosen target site. The first transformation into a Sm-R strain used the cassette to tag a target gene on the chromosome by homologous recombination, while conferring Kn-R but Sm-S on the recombinant. Replacement of the cassette by an arbitrary segment of DNA during a second transformation restored Sm-R (and Kn-S), allowing construction of silent mutations and deletions or other gene 31 replacements which lack a selectable phenotype. It was also shown that gene conversion occurred between the two rpsL alleles, in a process that depended on recA and that was susceptible to correction by mismatch repair. The Novel Conjugative Transposon, Tn1207.3 carries the Macrolide Efflux gene mef(A) in Streptococcus spp. Maria Santagati1, Francesco Iannelli2, Carmela Cascone1, Floriana Campanile1, Marco R. Oggioni2, Stefania Stefani1, Gianni Pozzi2. 1 University of Catania, Italy; 2 LA.M.M.B., Dipartimento di Biologia Molecolare, Università di Siena. The macrolide efflux gene, mef(A), confers the M-type resistance to macrolides and is found in different gram-positive genera, including Streptococcus, Enterococcus, Corynebacterium and Micrococcus. The 7,244 bp chromosomal element, Tn1207.1 was previously reported to carry the mef(A) gene in a clinical strain of S.pneumoniae. In this study, we investigated the presence of genetic elements homologous to Tn1207.1 in a clinical isolate of S.pyogenes displaying the M phenotype and carrying the mef(A) gene. In Streptococcus pyogenes 2812A, mef(A) was found to be carried by a 52-kb chromosomal genetic element which could be tansferred by conjugation. This genetic element was called Tn1207.3. DNA sequencing of the DNA adjacent to mef(A) showed that the 7,244 bp at the left end of Tn1207.3 was identical to Tn1207.1 whereas the overall size of Tn1207.3 was 52 kb, as established by pulsed-field gel electrophoresis. Tn1207.3-like genetic elements were found to be inserted at a single specific chromosomal site in 12 different clinical isolated of mef(A) carrying S.pyogenes. Tn1207.3 was transferred from S.pyogenes 2812A to Streptococcus pneumoniae by conjugation and DNA sequence analysis of six independent transconjugants showed that insertion of Tn1207.3 in the pneumococcal chromosome always occurred at a single specific site, which was identical to the insertion site of Tn1207.1. Using S.pneumoniae transconjugant MF2 as a donor, Tn1207.3 was transferred again by conjugation to S.pyogenes and S.gordonii. The previously described non conjugative element Tn1207.1 of S.pneumoniae appears to be a defective element, part of a longer conjugative transposon which carries mef(A) and is found in clinical isolates of S.pyogenes. 32