KC456067
advertisement

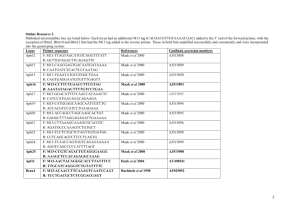
Electronic supplementary material: The behavioral and genetic mating system of the sand tiger shark, Carcharias taurus, an intrauterine cannibal. Demian D. Chapman1*, Sabine P. Wintner2, Debra L. Abercrombie1, Jimiane Ashe1, Andrea M. Bernard3, Mahmood S. Shivji3, Kevin A. Feldheim4. 1 School of Marine and Atmospheric Science & Institute for Ocean Conservation Science, Stony Brook University, Stony Brook, NY 11794, USA. 2 Kwa-Zulu Natal Sharks Board and Biomedical Resource Unit, University of KwaZulu-Natal, Durban 4056, South Africa. 3 Guy Harvey Research Institute & Oceanographic Center, Nova Southeastern University, Dania Beach, FL 33004, USA. 4 Pritzker Laboratory for Molecular Systematics and Evolution, Field Museum of Natural History, 1400 South Lake Shore Drive, Chicago, IL 60605, USA. * Author to whom correspondence should be addressed (demian.chapman@stonybrook.edu) Laboratory analysis Genomic DNA (gDNA) was isolated from a 0.25 mg sub-sample of tissue using the DNeasy Blood and Tissue DNA Isolation Kit (Qiagen, Valencia CA), following the manufacturer’s instructions. Ten microsatellite markers were amplified in the 15 mother-litter groups and 23 additional adults collected from Richard’s Bay. Five of these loci (Iox-12, CT6, Ct-109, Ct-69, Ct-93) were previously published [1]. These loci exhibited from 9-15 alleles and observed heterozity in Richard’s Bay from 0.667-1.0 (N=23 individuals). Five additional loci were developed using an enrichment protocol [2] PCR reactions for these new loci consisted of 1X PCR Buffer (10mM Tris-HCl, 50mM KCl, pH 8.3), 0.12 mM each dNTP, 10x BSA, 1.5 mM MgCl2 (Ct182 and Ct417) or 2.5 mM MgCl2 (Ct243, Ct432, Ct655), 0.16 μM of a fluorescently labeled universal M13 primer and the species-specific reverse primer and 0.04 μM of the species-specific forward primer with a 5’-M13 tail [3], 1 Unit Taq and approximately 40 ng genomic DNA. Thermal cycling profiles for Ct243, Ct417, and Ct432 were as follows: 94°C for 4min, followed by 37 cycles of 94°C for 15s, Ta (Table ESM1) for 15s and 72°C for 45s with a final extension of 72°C for 10min. Thermal cycling for Ct182 and Ct655 were as follows: 94°C for 4min, followed by 29 cycles of 94°C for 15s, Ta (Table ESM1) for 15s and 72°C for 45s followed by 7 cycles of 94°C for 15s, 53°C for 15s, 72°C for 45s with a final extension of 72°C for 10min. PCR products were mixed with LIZ-600 size standard (Applied Biosystems, Foster City, CA) and run on an ABI3730 DNA Analyzer. Scoring was verified by 2 or more experienced analysts (DC, JA, AB or KF) using Genemapper version 3.7 (Applied Biosystems). Locus Genbank Accession No. Ct182 Primer Sequence (5'-3') (forward first) Repeat motif Ta (oC) Allele size range (bp) N K HO HE TGTACGTGAGAACCCCAAGA (TATC)14..(TCTG)7 55 409-556 93 32 0.914 0.957 KC456065 CATTATATTGTATACATAATGGGTAG Ct243 GGTTTAGGAGGTGGGGTGTT (TAGA)16 48 229-261 119 9 0.874 0.859 KC456066 ACCCACTACCCTCTGCGTAA Ct417 CCTTCTCAAGCCCTCTAGCC (TAGA)27..(AGAC)8 53 331-453 83 23 0.880 0.915 KC456067 AGGAACAGAAAGGAGCCACA Ct432 AAATGCTAATAGGTAGATATTGATGGA (TAGA)26 53 278-398 84 20 0.821 0.912 KC456068 TTTTGAATAAGAGCTGAGAACCAA Ct655 TGCTTGTAATATCTGTCTGTCTGC (TATC)15 60 175-227 122 15 0.803 0.876 KC456069 GGCTTAGCCATATCCCATGA Table ESM1: New microsatellite loci developed for this study. Ta=annelaing temperature, N=sample size, K=number of alleles, HO = Observed heterozygosity, HE = Expected heterozygosity. Literature cited [1] Feldheim, K.A., Stow AJ, Ahonen, H., Chapman, D.D., Shivji, M.S., Peddemors, V., Wintner., S. 2007. Polymorphic microsatellite markers for studies of the conservation and reproductive genetics of imperilled sand tiger sharks (Carcharias taurus). Molecular Ecology Notes 7 (6), 1366-1368. [2] Glenn, T.C., Schable, N.A. 2005. Isolating microsatellite DNA loci. Methods in Enzymology. 395, 202-222. [3] Schuelke, M. 2000. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology 18, 233-234. Appendix I: Microsatellite genotypes of adult sand tigers used in this study Appendix II: Genotypes of embryonic sand tigers used in this study.