NUS researchers revealed three distinct structural reorganizations of
advertisement
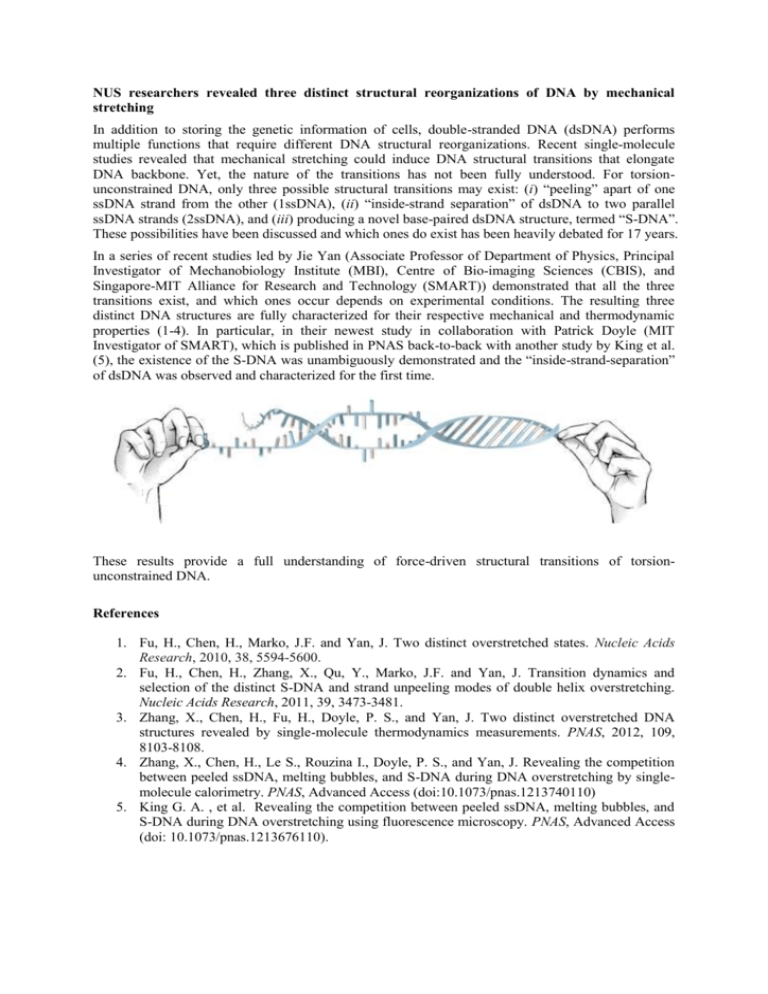
NUS researchers revealed three distinct structural reorganizations of DNA by mechanical stretching In addition to storing the genetic information of cells, double-stranded DNA (dsDNA) performs multiple functions that require different DNA structural reorganizations. Recent single-molecule studies revealed that mechanical stretching could induce DNA structural transitions that elongate DNA backbone. Yet, the nature of the transitions has not been fully understood. For torsionunconstrained DNA, only three possible structural transitions may exist: (i) “peeling” apart of one ssDNA strand from the other (1ssDNA), (ii) “inside-strand separation” of dsDNA to two parallel ssDNA strands (2ssDNA), and (iii) producing a novel base-paired dsDNA structure, termed “S-DNA”. These possibilities have been discussed and which ones do exist has been heavily debated for 17 years. In a series of recent studies led by Jie Yan (Associate Professor of Department of Physics, Principal Investigator of Mechanobiology Institute (MBI), Centre of Bio-imaging Sciences (CBIS), and Singapore-MIT Alliance for Research and Technology (SMART)) demonstrated that all the three transitions exist, and which ones occur depends on experimental conditions. The resulting three distinct DNA structures are fully characterized for their respective mechanical and thermodynamic properties (1-4). In particular, in their newest study in collaboration with Patrick Doyle (MIT Investigator of SMART), which is published in PNAS back-to-back with another study by King et al. (5), the existence of the S-DNA was unambiguously demonstrated and the “inside-strand-separation” of dsDNA was observed and characterized for the first time. These results provide a full understanding of force-driven structural transitions of torsionunconstrained DNA. References 1. Fu, H., Chen, H., Marko, J.F. and Yan, J. Two distinct overstretched states. Nucleic Acids Research, 2010, 38, 5594-5600. 2. Fu, H., Chen, H., Zhang, X., Qu, Y., Marko, J.F. and Yan, J. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Research, 2011, 39, 3473-3481. 3. Zhang, X., Chen, H., Fu, H., Doyle, P. S., and Yan, J. Two distinct overstretched DNA structures revealed by single-molecule thermodynamics measurements. PNAS, 2012, 109, 8103-8108. 4. Zhang, X., Chen, H., Le S., Rouzina I., Doyle, P. S., and Yan, J. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by singlemolecule calorimetry. PNAS, Advanced Access (doi:10.1073/pnas.1213740110) 5. King G. A. , et al. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy. PNAS, Advanced Access (doi: 10.1073/pnas.1213676110).