A Process for Microbial Hydrocarbon Synthesis
advertisement

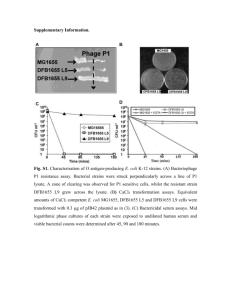
A Process for Microbial Hydrocarbon Synthesis: Overproduction of Fatty Acids in Escherichia coli and Catalytic Conversion to Alkanes Supplementary Material Rebecca M. Lennen1,2, Drew J. Braden1,2, Ryan A. West1,2, James A. Dumesic1,2, Brian F. Pfleger*1,2 1 Department of Chemical and Biological Engineering, University of Wisconsin-Madison 2 U.S. Department of Energy Great Lakes Bioenergy Research Center *Corresponding author: Brian F. Pfleger 3629 Engineering Hall 1415 Engineering Drive Madison, WI 53706-1691 phone: (608) 890-1940 fax: (608) 262-5434 pfleger@engr.wisc.edu Running title: Fatty acid overproduction and conversion to alkanes Sequence of synthesized BTE gene The full sequence of synthesized DNA includes restriction sites at the 5' (XmaI) and 3' (HindIII) ends (italicized), an artificial ribosome binding site (shown in blue), a spacer sequence (shown in red), an artificial start codon (bolded), and two bases following the start codon and before the XbaI site to generate an in-frame sequence (shown in green) is shown below. The codon corresponding to histidine-204 is underlined. The stop codon is bolded. 1 51 101 151 201 251 301 351 401 451 501 551 601 651 701 751 801 851 901 CCCGGGAGGA CTGCCTCAAC TCGTACTTTC CCATCCTGGC AAATCTGTTG TAAACGTGAC GCTACCCTAC TCCGGTAACA GGGCGAAATC GCACTCGTCG CCTGCTTTCA GCAAAAACTG CGCGCTGGAA GTTGCTTGGG CATTTCCTCT TTCTGCGCAG GTCTGCGACC GCGTACGGAG TAATTCCGGC GGATTATAAA TGCTGGATGA GCAATTCGTT CGTCATGAAC GTATCCTGGG CTGATGTGGG TTGGGGTGAC ATGGTATGCG CTGACGCGTT CCTGTCTACC TCGATAACGT AACGACTCCA CGACCTGGAT TCTTCGAGAC TTTACTCTGG CCTGACCACC ATCTGCTGCA TGGCGTCCAA GGAACCTCGT ATGACTCTAG TCACTTCGGT CTTATGAAGT CACATGCAGG TGATGGTTTC TAGTGCGTCG ACTGTGGAAG TCGCGATTTT GCACCTCCCT ATCCCGGACG GGCAGTTAAA CCGCGGACTA GTTAATCAGC TGTGCCGGAC AGTACCGTCG GTAAGCGGCG ACTGGAAGGC AGCTGACGGA GTTTAAGCTT AGTGGAAACC CTGCACGGTC GGGTCCAGAT AAGCCACCCT GGCACTACTC CACCCACGTA TCGAGTGTTG CTGGTCCGTG GAGCGTTCTG AAGTGCGCGG GACGACGAAA CATCCAGGGC ATGTGAACAA AGCATTTTCG CGAATGTACT GTTCTAGCGA GGCTCCGAAG TTCTTTCCGC GAAACCAAAA TGGTGTTTCG CGTTCTACCT GAATCACGCG TGGAAATGTC GCAGTAGAGC GATTGGCGCG ACTGTAAAAC ATGAACACCC TGAGATCGGT TCAAGAAACT GGTCTGACTC CCTGAAATAC AAAGCCATCA CGCGACTCCG GGCAGGTCTG TCCTGCGTGC GGCATCTCCG MG1655 MG1655 K27 MG1655 ΔfadD A glycerol + sodium oleate K27 MG1655 ΔfadD MG1655 ΔfadD ΔaraBAD sodium oleate B glycerol + L-arabinose L-arabinose Figure S1. Functional verification of gene deletions constructed in this study. (A) E. coli K-12 MG1655, K27 (a non-functional fadD mutant (Overath et al., 1969)), and MG1655 ΔfadD streaked on M9 minimal medium agar containing 0.4% glycerol and 0.1% sodium oleate (left), or 0.1% sodium oleate (right). K27 and the ΔfadD strain cannot grow on the oleate-only plate, indicating shut-down of β-oxidation. (B) K-12 MG1655 and MG1655 ΔfadD ΔaraBAD (RL08) streaked on M9 minimal medium agar containing 0.4% glycerol and 0.4% L-arabinose (left), and 0.4% L-arabinose (right). The ΔaraBAD strain cannot grow on the arabinose-only plate. % 57 100.0 43 75.0 71 50.0 85 25.0 99 0.0 50.0 75.0 100.0 112 126 125.0 141 156 150.0 184 175.0 210 221 200.0 225.0 255 250.0 288 275.0 300.0 319 325.0 Figure S2. Mass spectrum of tridecane in a decarboxylated decane extraction of the 34 hour culture of RL08/pBAD33-ACC/pBAD35-BTE. The molecular ion (m/z 184) is present. Figure S3. Representative mass spectra of various fatty acid methyl esters (10:1, top; 12:1, second from top; 12:0-OH, second from bottom; 14:1, bottom) not present in the standard mixture. Spectra are from a 23 hour extraction of a strain expressing BTE on plasmid pBAD35-BTE. The molecular ions for 12:1 (m/z 212), 12:0-OH (m/z 229), and 14:1 (m/z 240) are all present. Table SI: Molar concentrations of fatty acid methyl esters in fatty acid overproducing and negative control cultures prior to decane extraction, and alkanes following decane extraction and catalytic conversion. Species: Fatty acid methyl ester concentration (µmol mL-1 culture volume) 12:0 12:1 14:0 14:1 16:0 16:1 pBAD33-ACC/ pBAD18-BTE 1.44 0.11 0.23 0.01 0.21 0.00 0.21 0.01 0.22 0.00 0.10 0.00 pBAD33/ pBAD18-BTEH204A 0.01 0.00 ND* 0.05 0.00 ND* 0.24 0.01 0.02 0.00 Species: Alkane concentration (µmol mL-1 extract volume) undecane tridecane pentadecane pBAD33-ACC/ pBAD18-BTE 27.9 1.8 5.1 0.5 2.6 0.0 pBAD33/ pBAD18-BTEH204A 3.3 0.2 0.8 0.1 0.9 0.1 *ND = not detectable ACC (+) BTE (+) 8:0 10:0 10:1 12:0 15:0 14:1 12:0-OH 12:1 (std) 16:0 14:0 16:1 17:0 intensity (std) 18:1 18:0 ACC (-) BTE (+) ACC (+) BTE (-) ACC (-) BTE (-) standard mixture 3 6 9 12 15 18 21 24 time (min) Figure S4. Representative chromatograms (from 34 hour samples, undiluted hexane extracts) of strains in this study, with quantified peaks indicated. Very large increases in the levels of C12 and C14 species are clearly observed in BTE-expressing cultures. Labelled peaks that are not present in the standard mixture (10:1, 12:1, 12:0-OH, and 14:1) were identified by comparing their mass spectra to the NIST library.