1 - Exploring Weka`s interfaces, and working with big data
advertisement

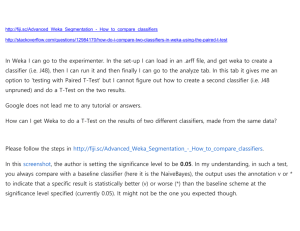
1 - Exploring Weka's interfaces, and working with big data Lesson 1.2 Activity: Percentage split vs cross-validation estimates 1. In the lesson video, Ian obtained the following results from J48 on the segment-challenge dataset, using 10 repetitions in both cases: mean std dev 10-fold cross validation 95.71% 1.85% 90% split 95.22% 1.20% (Note: 10 repetitions of 10-fold cross-validation involves executing J48 100 times.) Which do you think is a better estimate of the mean? 95.71% 95.22% Don't know 2a. In the above table it's curious that the standard deviation estimate for percentage split is smaller than that for cross-validation. Re-run the experiment and determine the standard deviation estimate for a 95% split, repeated 10 times. Answer: 2b. What is the standard deviation estimate for a 20-fold cross-validation, repeated 10 times? Answer: 2c. What is the standard deviation estimate for a 80% split, repeated 10 times? Answer: 2d. What is the standard deviation estimate for a 5-fold cross-validation, repeated 10 times? Answer: 3. Can you think of a reason why the standard deviation estimate tends to be smaller for percentage split than for the corresponding crossvalidation? The estimate is made using a different number of samples in each case. There's some overlap in the test sets for percentage split, but none for cross-validation. There's no reason; it's just a coincidence. Lesson 1.3 Activity: Comparing classifiers In the Experimenter, use J48, OneR and ZeroR (with default parameters) on the iris, breast-cancer, creditg, diabetes, glass,ionosphere, and segment-challenge datasets to get the following output. (Ian does exactly this in the lesson video, and the procedure is documented on the slides. Don't worry about the order of the rows, or the order of the columns -- so long as J48 comes first.) Now delete the segment-challenge dataset (which will make things much faster), add the NaiveBayes, IBk, Logistic, SMO, and AdaBoostM1 classifiers (all with default parameters), and perform the same test. From the output obtained, answer the following questions. 1. For one (and only one) of the datasets, some schemes significantly outperform J48 (at the 5% level). Which dataset? ionosphere credit-g iris breast-cancer glass diabetes 2. One of the classifiers is not significantly different from J48 on any dataset. Which classifier? AdaBoostM1 NaiveBayes Logistic SMO IBk 3. One of the classifiers is significantly better than J48 on exactly one dataset, and significantly worse than J48 on exactly one other dataset. Which classifier? AdaBoostM1 NaiveBayes Logistic SMO IBk 4. Change the analysis parameters to compare the various classifiers with OneR instead of with J48. There's one dataset on which all classifiers significantly outperform OneR. Which dataset? ionosphere glass diabetes breast-cancer credit-g iris 5. Which classifier significantly outperforms OneR on the largest number of datasets? AdaBoostM1 NaiveBayes Logistic SMO IBk 6. Change the analysis parameters to compare the various classifiers with SMO instead of with OneR. Which classifier significantly outperforms SMO on the largest number of datasets? AdaBoostM1 NaiveBayes Logistic J48 IBk 7. Which other classifier significantly outperforms SMO on at least one dataset? IBk NaiveBayes Logistic SMO AdaBoostM1 8. Ignoring whether or not the differences are statistically significant, which classifier performs best of all on each of the following datasets: a) Best performing classifier for iris: J48 IBk NaiveBayes Logistic SMO AdaBoostM1 b) Best performing classifier for breast-cancer: J48 IBk NaiveBayes Logistic SMO AdaBoostM1 c) Best performing classifier for credit-g: J48 IBk NaiveBayes Logistic SMO AdaBoostM1 d) Best performing classifier for diabetes: J48 IBk NaiveBayes Logistic SMO AdaBoostM1 e) Best performing classifier for glass: J48 IBk NaiveBayes Logistic SMO AdaBoostM1 f) Best performing classifier for ionosphere: J48 IBk NaiveBayes Logistic SMO AdaBoostM1 Lesson 1.4 Activity: Looking inside cross-validation Let's look at the models produced during cross-validation. To begin, open the Knowledge Flow interface and recreate the first example given in the lesson video. Use an ArffLoader; right-click it and select Configure to set the file iris. Connect the ArffLoader to a ClassAssigner, then make a dataSet connection to a CrossValidationFoldMaker, then make a trainingSet and a testSet connection to J48, then make a batchClassifier connection to a ClassifierPerformanceEvaluator, then make a text connection to a TextViewer. Add a GraphViewer and connect the graph output of J48 to it. Run the system (Start loading on the ArffLoader right-click menu) and examine the results of the GraphViewer. This shows the 10 models that are produced inside the cross-validation: click to see each one. It's easier to compare them using a TextViewer rather than a GraphViewer, so connect a TextViewer up to J48 and use it to look at the models (you'll need to run the system again). 1. Among the 10 models there are two basically different structures. How many leaves do they have respectively? (Type your answer as two integers separated by a space.) Answer: 2. How many of the 10 models have the larger number of leaves? Answer: 3. The first of the numbers that appear in brackets after each leaf is the number of instances that reach that leaf. The total number of instances that reach a leaf is the same for each of the 10 trees. What is that number of instances? 130 135 122 150 4. When two numbers appear in brackets after a leaf, the second number shows how many incorrectly classified instances reach that leaf. One of the 7 models with 5 leaves makes fewer misclassifications than the other 6 models. How many does it make? Answer: 5. The 3 models with 4 leaves all differ slightly from each other. How? They branch on different attributes. They branch on the same attributes but the cutoff values differ. They have a smaller number of class values. A different total number of instances reach the leaves. Lesson 1.5 Activity: Using Javadoc and the Simple CLI 1. Use Weka's Javadoc to find out what the "-I" (capital i, not lower-case l) option of the IBk classifier does. What does the documentation say about this? Weight neighbors by 1 - their distance. The nearest neighbor search algorithm to use. Weight neighbors by the inverse of their distance. Number of nearest neighbors (k) used in classification. 2. In the Explorer, configure IBk to use this option (by setting distanceWeighting to Weight by 1/distance), set the KNN parameter to 5, click OK, right-click the configuration (that is, the large field that appears to the right of the Choose button), and copy it to the clipboard. Paste this command into the Simple CLI interface, preceded by "java" and followed by "-t iris.arff" (i.e., specify iris as a training file; you will have to precede it by an appropriate directory specification). Then press Enter to run the command. What is the percent accuracy of correctly classified instances? (Here and elsewhere in this course, round percentage accuracies to the nearest integer.) Answer: 3. In preparation for Lesson 1.6, use Weka's Javadoc to find out which classifiers are "updateable", i.e., implement the UpdateableClassifier interface. (Hint: IBk is one of them, and near the top of its "Class" page are links to all the interfaces it implements. Just click the appropriate one.) How many updateable classifiers are there? Answer: Lesson 1.6 Activity: Experience big data Warm-up exercise. Reproduce what Ian was doing in the Command Line interface with the LED24 data in the lesson: make a test file with 100,000 instances using java weka.datagenerators.classifiers.classification.LED24 -n 100000 -o make a training file with 10,000,000 instances using java weka.datagenerators.classifiers.classification.LED24 -n 10000000 -o apply NaiveBayesUpdateable: java weka.classifiers.bayes.NaiveBayesUpdateable -t -T (this may take a few minutes) verify that Weka runs out of memory if cross-validation is attempted: java weka.classifiers.bayes.NaiveBayesUpdateable -t if you feel particularly brave, repeat the exercise with a 100,000,000-instance training file Actual exercise. The covertype data has 581,012 instances, 55 attributes, and 7 class values. Working with big data is a pain, so we've created a small 10,000-instance subset, covtype.small, for you to play with. 1. Using the Explorer, determine the percentage accuracy of Naive Bayes, evaluated on the training set. 66% 65% 68% 67% 2. Using the Explorer, determine the percentage accuracy of NaiveBayes evaluated by cross-validation accuracy. 67% 68% 65% 66% 3. Using the Simple CLI, determine the percentage accuracy of NaiveBayesUpdateable evaluated on the training set. 68% 66% 67% 65% 4. Using the Simple CLI, determine the percentage accuracy of NaiveBayesUpdateable evaluated by cross-validation accuracy. 67% 65% 68% 66%