Phylogenetic Analysis Using BLAST, CLC, Clustal and MacClade
advertisement
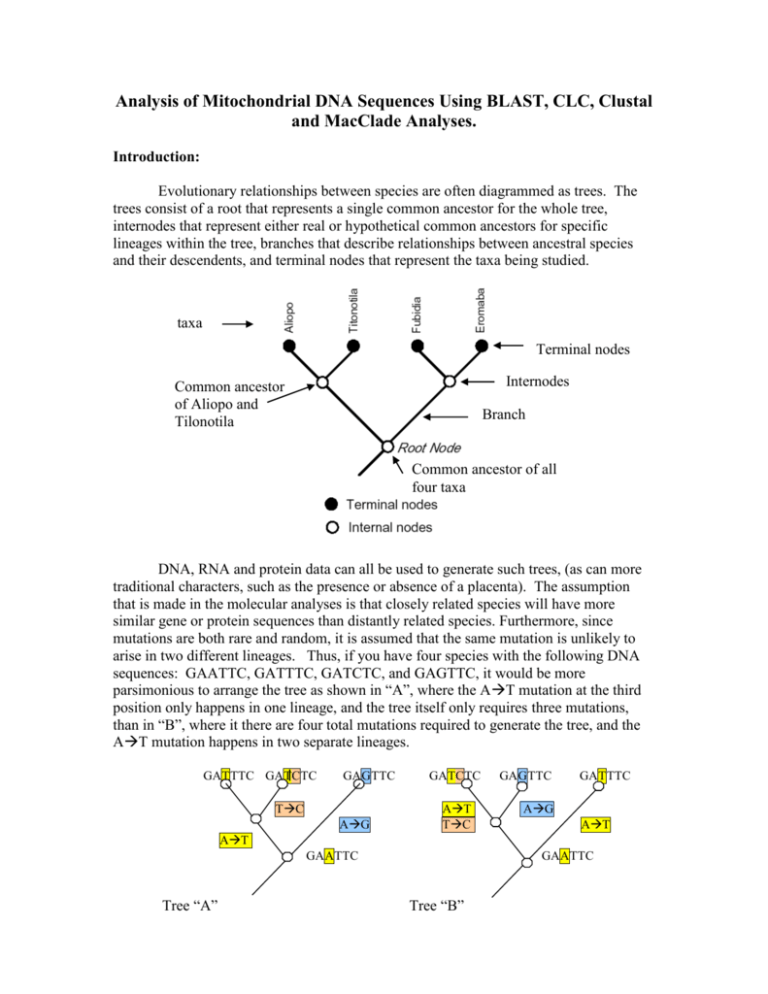
Analysis of Mitochondrial DNA Sequences Using BLAST, CLC, Clustal and MacClade Analyses. Introduction: Evolutionary relationships between species are often diagrammed as trees. The trees consist of a root that represents a single common ancestor for the whole tree, internodes that represent either real or hypothetical common ancestors for specific lineages within the tree, branches that describe relationships between ancestral species and their descendents, and terminal nodes that represent the taxa being studied. taxa Terminal nodes Internodes Common ancestor of Aliopo and Tilonotila Branch Common ancestor of all four taxa DNA, RNA and protein data can all be used to generate such trees, (as can more traditional characters, such as the presence or absence of a placenta). The assumption that is made in the molecular analyses is that closely related species will have more similar gene or protein sequences than distantly related species. Furthermore, since mutations are both rare and random, it is assumed that the same mutation is unlikely to arise in two different lineages. Thus, if you have four species with the following DNA sequences: GAATTC, GATTTC, GATCTC, and GAGTTC, it would be more parsimonious to arrange the tree as shown in “A”, where the AT mutation at the third position only happens in one lineage, and the tree itself only requires three mutations, than in “B”, where it there are four total mutations required to generate the tree, and the AT mutation happens in two separate lineages. GATTTC GATCTC GAGTTC TC AG GATCTC AT TC GAGTTC GATTTC AG AT AT GAATTC Tree “A” GAATTC Tree “B” Procedure: BLAST Analysis of Human Mitochondrial DNA Sequences. a) Download the Class Mitochondrial DNA Sequences as instructed by your professor and find your number. If you are not there, your sequence was not of sufficient quality to analyze. Do not worry, simply choose another sequence and click on the link. b) Select the entire sequence record, starting with > and ending with the last base of the sequence. That format is called 'FASTA', and is a common sequence format for almost all sequence analysis programs. Copy the sequence, and then go to the NCBI BLAST database at http://blast.ncbi.nlm.nih.gov/Blast.cgi. c) First, select nucleotide BLAST, as you did for your pre-lab activity, and paste your sequence in the BLAST window. Is your sequence human mitochondrial DNA? d) Next, go back to the BLAST home page, and scroll down to the 'Align two sequences (bl2seq)' link in the 'Special' window. Align your sequence with that of Rarely Reclusive. Is it a perfect match? Worksheet Question 1: Give the E value, the percent identity and the Score. Do you think that Rarely is closely related to this you? Why or why not? Phylogenetic Analysis - Creating Alignments and Trees Using UPGMA Analysis During this part of the activity, you will learn how to use the CLC Free Workbench to align DNA sequences and to generate a UPGMA tree. Now you are ready to align your sequences so that you can perform your phylogenetic analysis. Creating a sequence alignment makes sure that you are comparing homologous portions (beginnings to beginnings, and ends to ends) of all of the genes in the Genetics project. a) Open up the CLC Free Workbench program from your dock. Import the class FASTA sequences by clicking on the small import folder icon in the upper toolbar. b) In the Toolbox window (below the Navigation Area) double click on 'alignments and trees' and then on 'create alignment'. Click on 'class sequences' and then click on the right arrow to move the sequences to the right analysis window. Click on 'next'. c) Under Gap settings, set Gap Open Cost to 100. This will discourage the computer from trying to insert gaps in your sequences while matching them up. d) Under End Gap Cost choose 'Free'. Since our sequences differ in length because of the quality of your sequencing reactions (rather than a real evolutionary difference) this setting will keep the computer from paying too much attention to sequences that are longer or shorter than one another. e) Make sure that 'Fast Alignment' is checked, and click on 'Finish'. f) The computer will now align all of the sequences to each other (as opposed to simply aligning two sequences as you did in BLAST (bl2seq). The aligned sequences will open in a large window to the right of the Navigation Area. The computer also generates a consensus sequence for all of the data, which represents the most common sequence that all of the class sequences share. You can see the consensus sequence below the aligned class sequences. If you scroll up and down the alignment window, you can see that the matches are not perfect, and that the computer has inserted many gaps to make the sequences align. g) Now it is time to generate a phylogenetic tree from your aligned sequences. Under the toolbox, double click on Create Tree. In the Selected Elements box you should see 'mito124.txt_alignment'. Click on 'Next'. Under 'Algorithm' choose UPGMA to tell the computer to use that method to construct the tree. Make sure that the boostrap analysis is selected for at least 100 replicates. You can go up as high as 1000 replicates. h) Click on 'Finish'. i) You can now see the UPGMA Phylogenetic Tree in your main window. j) In the right window, under 'Tree Settings', choose 'Tree Layout' -> Node symbol - > 'None' and make sure 'Layout' is set to 'Topology' (the branch lengths will not be proportional to the UPGMA values as they would be if you choose 'Standard', but they will be easier to see). k) Under 'Annotation Layout' make sure that 'Branches' are set to length, rather than the bootstrap value. Worksheet Question 2: Which group members are most closely related to one another? Worksheet Question 3: Which group members are most closely related to Rarely Reclusive? l) Finally, you need to export your tree in .jpg format for your worksheet. Make sure your tree window is selected, and then click on 'Graphics' in the upper program bar. Make sure you have chosen JPG formatting from the drop down list. Make sure your file name has .jpg at the end. Choose to save the whole view in high resolution (the middle value), then click on 'export'. Save the file to your desktop. You should see something like this, when you open the picture file: Worksheet Question 4: Import your picture file into the appropriate space in the worksheet. Worksheet Question 5: Interpret this phylogenetic tree. Which one of your group members is most closely related to Rarely Reclusive? Worksheet Question 6: Which one of your classmates is most closely related to Rarely Reclusive? Worksheet Question 7: For each group member, state the person to whom they are most closely related, according the UPGMA analysis. Phylogenetic Analysis - Human Evolution and Your Place in the Human Family a) Well, you can't all be most closely related to Rarely, so who are you related to? You will now do Clustal Analysis to determine the answer to this question. b) Follow this link (http://www.bioservers.org/bioserver/) to the Dolan DNA Learning Center login page. Create a username, a password, and log in to the Sequence Server site. c) Click on 'Manage Groups' from the menu in the center of the top of the page. You will now see a window that looks like this: d) Click on the upper right pull-down menu (sequence sources). You should now see a variety of choices. Choose 'modern human mt DNA'. Click on the boxes to the left of each of the sequences to select them, and then click on 'ok'. e) Click on 'Manage Groups' again. This time select 'Prehistoric Human mt DNA' and click on all of the boxes, and then 'ok'. f) You have now uploaded a whole bunch of mtDNA sequences from the database for analysis. g) To upload your own sequence, go 'Manage Groups' one last time. This time select 'Public’ and click on ‘Susquehanna Genetics 2009’. Select your sequence, and the sequences of the members of your group, and mitorarely (Rarely Reclusive). h) To compare your group members to the available sequences, click on the box to the left of your sequences, and then next to any other sequences that interest you (you'll see lots of drop-down possibilities within each group). You may only select up to ten total sequences (including your group sequences). Have fun, but also use sequences that make sense, given what you know about your ancestry. i) After you have selected 10 sequences for analysis, find the word 'compare' in the gray bar menu, and choose 'phylogenetic tree' from the drop-down box. Click on "Compare". After your sequences are analyzed, a popup window will show you your tree. This tree is based on an alignment program called “Clustal”. Clustal Analysis generates a pairwise score for every pair of sequences that are to be aligned, much like we did with the CLC alignment program. These scores were calculated as the number of identities in the best alignment divided by the number of residues compared (gap positions are excluded). l) Choose 'phenogram' and 'yes' (to make the tree branch lengths proportional to the evolutionary distances). You will see something that looks like the picture below, but that contains the individuals and species that you chose to analyze. m) Phenograms (phylograms) such as this one can provide a ton of information. First of all, a phylogram is assumed to be an estimate of a phylogeny. IN other words, the branch lengths are proportional to the amount of inferred evolutionary change. A cladogram, on the other hand, is a branching diagram (tree) assumed to be an estimate of a phylogeny where the branches are of equal length. Therefore, cladograms show common ancestry, but do not indicate the amount of evolutionary "time" separating taxa. One thing that this phylogram shows is that Lake Mungo Man and African American #1 share a common ancestor, to which Lake Mungo Man is more closely related. Worksheet Question 8: Right click on the phenogram (command click on the Mac) and select ‘copy picture.’ Paste your phenogram into the appropriate space in the worksheet. Worksheet Question 9: Interpret your phenogram. Which human ancestral lineages is each of your group members most related to? Worksheet Question 10: Which one of your group members is the most closely related to Rarely Reclusive according to this Clustal-based analysis? Worksheet Question 11: Does this analysis agree with your CLC-based analysis? MacClade Analysis of Mitochondrial Sequences and Maximum Parsimony In this exercise you will use MacClade to generate cladograms or phylograms (you can choose) that have the smallest tree length (this is analogous to the total number of mutations needed to account for the tree) while minimizing the number of times that a single mutation arises in multiple lineages. You will then compare your MacClade tree to the ones you generated using the CLC/UPGMA and Clustal-based analyses. Procedure: Download the National Biomedical Research Foundation (NBRF) Format File from the syllabus (filename mitochondrial.NBRF) to your desktop by typing control+click on the file name. Open MacClade 1. Double Click on the MacClade Icon. 2. Choose ‘Open File’ (Your computer may open a Finder Window instead of allowing you to choose Open File..this simply takes you directly to step 3, so proceed!). 3. Open ‘mitochondrial.NBRF’ from your desktop. 4. Click ‘OK’ to verify that your file is an NBRF DNA file. 5. You should now see that the mitochondrial sequence files have been uploaded into the MacClade program as seen in the picture below: 6. While this window contains all of the sequences (the taxa in this case are the individual students, and the characters are the individual DNA bases at each position of the sequence), the sequences are not yet aligned, so we need to do that right after you save your data. Save Data 1. Under FILE choose SAVE FILE AS, and give your file a name and save it to the desktop. This will make sure you do not lose your data if something goes wrong during later manipulations. Don’t forget to throw away all of your files and to empty the trash before you put your computer away for the semester. Aligning DNA Sequences 1. In order to analyze how closely related these gene sequences are using MacClade, you need to make sure that each sequence is properly lined up. 2. MacClade’s Alignment Tool can be seen in the toolbox in the bottom left of your MacClade window, as indicated by the arrow below: 3. Click and hold on the Alignment Tool. In the popup box, select ‘slow method using less memory’. If you do not do this, the program will crash!!! 4. To align two sequences, make sure that the alignment tool is selected. Now, click on the lower sequence, and drag it up to the one directly above it. Release. The computer will now line up the two sequences so that they match as closely as possible. Save your data. I recommend you save after every alignment step below (sometimes this program crashes) 5. Repeat by dragging the third sequence up to the second, and so on, until you have finished aligning all of the sequences to the sequence above them in a pairwise fashion (yes it’s a pain to only align them pairwise…but that is all this program can do). 6. When you finish, you should be able to see (the bases are each colorcoded) that the sequences line up really nicely now. You are ready to generate a phylogenetic tree. Manipulate Data 1. Generate tree a) Under WINDOWS choose TREE WINDOW b) Choose DEFAULT LADDER - a tree should now appear! This tree makes no assumptions at all about how your microbes are related. In fact, if you look at the tree, it simply places the organisms in the same order in which you entered the sequences. c) Under TRACE choose TRACE ALL CHANGES. This will provide a color visualization of the number of nucleotide changes between a common ancestor and the next ancestor or terminal taxon. If you place the cursor arrow on a tree branch, the number of unambiguous nucleotide changes in that branch will be written in the small box on the bottom right. d) Under Display choose ‘Tree Shape and Size”. The choices, from left to right below are either an angled or a square branch cladogram, or a phylogram. 2. Minimize Tree a) Find the most parsimonious tree by clicking on branches, and then dragging them to new tree locations, remembering as you do so that you are changing the assumptions about how the species are related. Your goal is to search for the tree that gives the lowest number of “steps” (analogous to the total number of nucleotide changes necessary to account for the proposed evolutionary relationships - so lower is better). If you want to know what the theoretical smallest tree is, you can choose minimum possible from the upper program bar. Both the current tree length and the minimum possible tree length will be displayed in a small box in the lower right hand corner of your tree. Try to get as close to the minimum as possible. b) Try manipulating the tree so it looks like the ‘ideal’ tree from your Dolan DNA Center analysis. Have you found the minimum yet? c) Now that you’ve played around with the tree a bit, it is time to tell you a little secret – the computer will actually help find the smallest tree for you. Simply click on the ‘search above’ icon in the toolbox, and then move the cursor to the root of the tree. When you click on the root, the program will search for, and display, the shortest tree above that spot. d) Once you are convinced you have the smallest tree, save your file and then go to File>Save Graphics File. Choose either a phylogram or a cladogram from the left menu, and then under Options in the right drop down menu choose Legends and click on ‘trace legend’ and ‘tree statistics’. Under Options “Branch Shading” choose colors. Click on PICT File and save the file to your desktop. Worksheet Question 12: Import the PICT file into the appropriate spot in the worksheet. Worksheet Question 13: Interpret this phylogenetic tree. Which one of your group members is most closely related to Rarely Reclusive? Worksheet Question 14: Which one of your classmates is most closely related to Rarely Reclusive? Worksheet Question 15: For each group member, state the person to whom they are most closely related, according to the MacClade analysis. Worksheet Question 16: Does this analysis give you a different answer than the other two analyses did? If so, why do you think that is? Workshop Genetics Worksheet: Analysis of Mitochondrial DNA Sequences Using BLAST, CLC, Clustal and MacClade Analyses. Names__________________________________________________________________ Worksheet Question 1: Give the E value, the percent identity and the Score. Do you think that Rarely is closely related to this you? Why or why not? Worksheet Question 2: Which group members are most closely related to one another? Worksheet Question 3: Which group members are most closely related to Rarely Reclusive? Worksheet Question 4: Past your picture file here. Worksheet Question 5: Interpret this phylogenetic tree. Which one of your group members is most closely related to Rarely Reclusive? Worksheet Question 6: Which one of your classmates is most closely related to Rarely Reclusive? Worksheet Question 7: For each group member, state the classmate to whom they are most closely related, according the UPGMA analysis. Worksheet Question 8: Paste your phenogram here. Worksheet Question 9: Interpret your phenogram. Which human ancestral lineages is each of your group members most related to? Worksheet Question 10: Which one of your group members is the most closely related to Rarely Reclusive according to this Clustal-based analysis? Worksheet Question 11: Does this analysis agree with your CLC-based analysis? Worksheet Question 12: Import the PICT file here. Worksheet Question 13: Interpret this phylogenetic tree. Which one of your group members is most closely related to Rarely Reclusive? Worksheet Question 14: Which one of your classmates is most closely related to Rarely Reclusive? Worksheet Question 15: For each group member, state the person to whom they are most closely related, according the MacClade analysis. Worksheet Question 16: Does this analysis give you a different answer than the other two analyses did? If so, why do you think that is?