Supplementary Materials and Methods RNA extraction and
advertisement

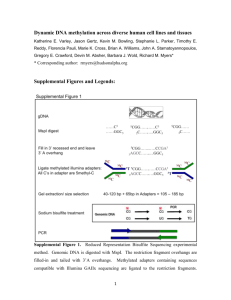
Supplementary Materials and Methods RNA extraction and quantitative reverse-transcription (qRT)-PCR Total RNA was extracted with TriPure Isolation Reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. qRT-PCR was performed using M-MLV reverse transcriptase (Promega, Madison, WI) and LightCycler® 480 SYBR Green I in the LightCycler® 480 system. The mRNA level was normalized to EF-1α (Elongation Factor 1- α), as we have described (Serre et al 2010). Fluorescence microscopy After serum/E2 starvation for 3 days, cells were fixed with 4% paraformaldehyde and permeabilized with methanol. Then cell were stained with anti-Cav1 (clone 2297) antibody (BD Biosciences, San Diego, CA) and counterstained with DAPI. The Applied Precision DeltaVision personalDV Live Cell Imaging System was used to visualize cells. Migration/invasion assays Cells were grown to 80% confluence and then serum starved overnight prior to use. Cells were washed with PBS, harvested using HyQTase solution (Thermo Scientific, Rockford, IL), and resuspended in starvation medium (MEM with 0.1% BSA). Transwell chambers (24-well; BD BioCoat Control Inserts from BD Biosciences) with 8.0-μm pore size polycarbonate membrane were used for migration assay, and BD BioCoat Matrigel Invasion Chamber (BD biosciences) was used for invasion assays. Cells were plated at a density of 5 × 104 per well in the upper well (except MDAMB231 cells, which were seeded at 1× 104 per well), which was placed into a lower well containing complete growth medium. After incubation for 24 h, cells were wiped from the interior of the insert. Cells that had migrated through the membrane to the bottom of the insert were stained using the Hema 3 staining system (Fisher Scientific, Pittsburg, PA) and observed under the optical microscope at a magnification of 100×. Cells were counted in 5 fields of triplicate membranes. Methylation specific PCR and bisulfite genomic DNA sequencing For detecting the methylation on the Cav1 CGI using MSP, 5’-GGTATTTTTGTAGGCGCGTC-3’ (sense) and 5’-CTAACAACAAAAAACGAAAAACG-3’ (antisense) were used for methylated form (producing a fragment of 210 bp with annealing at 55°C), and 5’-GTTTATATTGGGTATTTTTGTAGGTGTGT-3’ (sense) and 5’-TCCCCAAAATTCTAACAACAAAAAACAAAAAACA-3’ (antisense) were used for unmethylated form (producing a fragment of 231 bp with annealing at 58°C) (Lin et al 2004). For detecting Cav1 5’ CGI shore methylation using MSP, 5’-TTATTTCGAAGCGTTTGGGAG-3’ (sense) and 5’-AACACTCGTTTACATCTAATCG-3’ (antisense) primers were used for the methylated sequence, and 5’-TTATTTTGAAGTGTTTGGGAG-3’ (sense) and 5’-AACACTCATTTACATCTAATCA-3’ (antisense) were used for the unmethylated sequence (producing a fragment of 105 bp with annealing at 57°C) (Sunaga et al 2004). Methylated and unmethylated control DNAs were purchased from QIAGEN (Valencia, CA). For bisulfite genomic DNA sequencing, 5’-ATAGGGTAGGATTGTGGATTGT-3’ (sense) and 5’-TAAACACATCCCCAAAATTCTAAC-3’ (antisense) were used to amplify (with annealing at 55°C) the region located at 5’ of Cav1 CGI (Sunaga et al 2004). We evaluated the methylation status as described previously (Sunaga et al 2004). Bisulfite treatment converts unmethylated C to T but does not alter methylated C. If only a T was detected at this position in sequencing, the status of this CpG dinucleotide was defined as “fully unmethylated” (open square). If only C was detected, this CpG dinucleotide was defined as “fully methylated” (filled square). Detection of both T and C and equal peak heights were defined as 50% methylation (50% filled square). If the C peak was higher than the T peak, methylation was defined as 75%. If the C peak was smaller than T peak, methylation defined as 25%. MBDCap-seq and data analysis Methylated DNA was eluted by the MethylMiner Methylated DNA Enrichment Kit (Invitrogen) according to the manufacturer's instructions. Methyl-CpG binding domain-based capture (MDBCap) libraries for sequencing were prepared following standard protocols from Illumina (Illumina Inc., San Diego, CA). MBDCap coupled with massively parallel sequencing (MBDCap-seq) libraries were sequenced using the Illumina Genome Analyzer II (GA II) as per manufacturer's instructions. Sequencing was done up to 36 cycles for mapping to the human genome reference sequence. Image analysis and base calling were carried out with the standard Illumina pipeline. Differentially methylated regions were quantified for pairwise comparison by the difference of methylation values. RNA-seq Total RNA was purified from MCF7, MCF7-T and MCF7-F cell lines with Trizol (Invitrogen) and then further purified with the RNeasy Plus mini kit (Qiagen) to remove all traces of DNA and protein. RNA samples were subjected to limited fragmentation (Ambion) followed by purification using the RNeasy MinElute kit (Qiagen). Orientation-specific smRNA adapters (Illumina v1.5) were ligated sequentially with T4 RNA ligase using a two-step process. Ligated RNA fragments were then purified with RNeasy MinElute and converted to double stranded DNA with SuperScript III reverse transcriptase (Invitrogen) followed by RNaseH treatment and PCR amplification with Phusion DNA polymerase (NEB, Ipswich, MA). These primary RNA-seq libraries were purified with Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN) and quantified using either the Nanodrop spectrophotometer (Thermo) or Qubit fluorometer (Invitrogen), prior to ribosomal RNA reduction with our optimized protocol for DSN normalization (Evrogen). Following DSN normalization and AMPure XP bead re-purification, RNA-seq libraries were again amplified by PCR with an additional AMPure XP bead purification. These finished libraries were characterized with the Qubit fluorometer and Agilent Bioanalyzer high-sensitivity DNA kit for quantification along with RT-PCR to validate rRNA reduction. ChIP-seq MCF7 and MCF7-T cells were cross-linked with 1% formaldehyde and immunoprecipitated with antibodies against H3K4me2, H3K9me2 (Abcam, Cambridge, MA), and H3K27me3 (Diagenode, Denville, NJ), as described previously (Feng et al 2008). After purification, “ChIPed” DNA samples were separated by 12% PAGE and the 100-300 bp DNA fractions were excised and eluted from gel slices. Then, Illumina library was constructed and sequenced using an Illumina Genome Analyzer. Feng W, Liu Y, Wu J, Nephew KP, Huang TH, Li L (2008). A Poisson mixture model to identify changes in RNA polymerase II binding quantity using high-throughput sequencing technology. BMC Genomics 9 Suppl 2: S23. Lin SY, Yeh KT, Chen WT, Chen HC, Chen ST, Chang JG (2004). Promoter CpG methylation of caveolin-1 in sporadic colorectal cancer. Anticancer Res 24: 1645-1650. Serre D, Lee BH, Ting AH (2010). MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 38: 391-399. Sunaga N, Miyajima K, Suzuki M, Sato M, White MA, Ramirez RD et al (2004). Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res 64: 4277-4285.