Nucleotides
advertisement

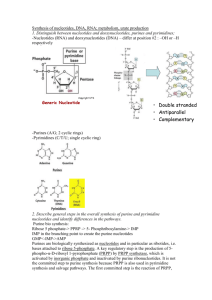
Nucleotides-Introduction and Purines 10/15/2008 6:14:00 AM Why study? Genetic disorders of nucleotide metabolsm cause significant and serious diseases. Cancer cells show an increased production of nucleotides. Inhibiting nucleotide synthesis inhibits growth. Using this, we can develop drugs for cancer, antibiotics. Cancer cells are more sensitive to nucleotide synthesis inhibitors than normal cells. Roles of Nucleotides Nucleotides are the activated precursers for making DNA and RNA (the genetic material) This makes them prime targets for arresting growth of cells. Activated intermediates in biosynthetic pathways. ATP, CDPglucose, S-adenosyl methionine ATP is used as energy currency Adenine is part of many coenzymes Metabolic regulation: Cyclic AMP Structure A nucleotide has three parts. It has a nitrogenous base (purine or pyrimidine), a sugar, and 1, 2 or 3 more phosphates. You must have all three parts to have a nucleotide. A nucleoside does not have the phosphates (adenosine, uracil, etc) Three types of bonds The base is attached to the suger by an N glycosidic bond The first phosphate is contected to the sugar by a phosphate monoester (ester is an acid to an alcohol The phosphates are bound together by acid anhydride bonds (this is the high energy bond that is split to give 7 kcal/mole) 1 The Bases Two flavors: Pyrimidine (one ring) and purine (two ring) Heterocyclic (carbon and nitrogen are both involved in the ring Aromatic and planar Pyrimidines number in a clockwise fashion starting at the bottom o 1 Nitrogen attached to the sugar o One urea unit (NCN) and one CCC unit o Cytosine (two additions to ring), Thymine (three additions to ring), Uracil (two additions) Purines start on the 6 member ring and number 1-6 in a counter clockwise fashion, then number the other 3 atoms on the 5 member ring 7, 8, 9 o Sugar is bound at the 9 nitrogen o One CCC unit sandwiched by two urea units. o Adenine (one addtion) and Guanine (two additions) The sugar If the sugar is ribose, the nucleoside is called a ribonucleoside If the sugar is deoxyribose, the nucleoside is a deoxyribonucleoside Five carbon is outside the ring DNA and RNA are polymers of nucleosides A:T 2 G:C Nucleotides names Usually longer than the bases name itself. Apparently that should help us remember them. AdenineAdenosine CytosineCytodine ThymineThymidine****** GuanineGuanidine UracilUridine ******Because Thymidine is almost exclusively found in DNA, it is understood that Thymidine is deoxythymidine, unless otherwise stated. Nucleotide names Add Nucleoside ________phosphate (fill in blank with mono, di, tri) Odd Ball Nucleotides Intermediates in some of the pathways Hypoxanthine (Purine-one addition) (nucleotide is called inosine monophosphate because scientists are lazy and wont say hypoxanthine monophosphate) Xanthine (Purine-two additions) Purine Synthesis Although purines and pyrimidines are often available in the diet, the body likes to make them de novo (from scratch) Two sources of ribose-5-phosphate o Oxidative (glucose-6-phosphate dehydrogeogenase) forward PPP) o Non oxidative using G3P and F6P (backwards PPP) 3 1. First step is PRPP—puts a pyrophosphate on the 1 carbon. This is an energy using step that requires ATP. 2. Second step (first committed step)—PRPP Glutamyl Amidotransferase Glutamine is an amino donor. The step cleaves the pyrophosphate and replaces it with an Amino. Uses water and glutamine. Adds the N9 3. Third Step—glycine is added and ATP is hydrolyzed and C4, C5 and N7 4. Formyl group from THF (Formyl Transferase) N10FormylTetrahydrofolate is used and C8 is added 5. N3 is added. Glutamine is the donor 6. Five member ring closure 7. C6 is added by carboxylation 8. Aspartate is added 9. Fumarate is cleaved leaving N1 10. C2 is added by N10Formyl-Tetrahydrofolate 11. Ring Closure—First purine ring formed is Inosine Monophosphate The next steps differ in the production of Adenosine Monophosphate and Guanisine Monophosphate o AMP SynthesisAspartate serves as an amino donor to make adenosine monophosphate ( two steps: add the entire aspartate then cleave it off leaving the amino) o GMP synthesisIMP Dehydrogenase to make Xanthosine monopphospate and then glutamine is used as an amino group donor that replaces the carbonyl added by IMP dehydrogenase o Regulation of synthesis Negative feed back. First and second step are inhibited by IMP , AMP and GMP PRPP is not purely for Purine synthesis IMPwhatever step is regulated by its own end produce AMP shuts down AMP synthesis GMP shuts down GMP synthesis 4 Purine Synthesis Overview Purine Ring is built into a preexisting ribose-5-monophosphate (deoxyribose-5-monophosphate) Ring atoms are added one at time with the exception of step 3 with adds an entire glycine Glutamine serves as an amino group donor in steps 2, 5 and 15 Formyl Tetrahydrofolate serves as carbon donor in steps 4 and 10 The first nucleotide formed is IMP, which is then converted to AMP or GMP by branched pathways Glutamine Rxn 2 (N9) Rxn 5 (N3) Rxn 15 (Guanosine) Aspartate Rxn 8 (N1) Rxn 12 (Adenosine) N10 Tetrahydrofolate Rxn 4 (C8) Rxn 10 (C2) Glycine Rxn 3 (C4, C5, N7) Carbon Dioxide Rxn 7 (C6) NADH Produced Step 14 (Guanosine) ATP Consumed Rxn 1-PRPP Rxn 3 Catabolism of Purines Adenosine 1. Demamination of C6 (conversion of NH2carbonyl) releases ammonium and utilizes water. Enzyme is Adenosine Deaminiase. Deficiency in Adenosine Deaminase is involved with SCID 2. Purine Nucleoside phosphorylase. Same type of reaction as glycogen phosphorylase. Removes Ribose 1 phosphate 3. Xanthine Oxidase makes uric acid Guanosine 1. Guanosine uses Guanase to make xanthine 2. Xanthine Oxidase makes uric acid. 3. Primates stop here- Uric acid may have some antioxidant properties 5 CLINICAL SIGNIFICANCE Xanthinuria Genetic deficiency of Xanthine Oxidase. Causes increased secreastion of xanthine and hypoxanthine and a decrease in uric acid. May cause xanthine lithiasis (kidney stones) Gout-Hyperuricemia Hyperuricemia is the excess of uric acid in the blood or urine Can crystallize in the joints. Inflammed joints due to depsosition of uric acid crystals is called gout There are some genetic causes o PRPP sythetase mutation that is not subject to feed back regulation. If no feeback you make to many purines which leads to increased purine degredation o Glucose-6-phopshatse deficiency, increased glucose 6 phosphate leads to increased ribose 5 phosphate via PPP o Partial HGPRTase deficiency (purine salvage pathway) Treatment o Anti-inflammatory Colchecine Alkaloid that inhibits microtubule assembly in leukocytes (inflammatory response) NSAIDs o Inhibit Uric Acid Production Allopurinol inhibits Xanthine Oxidase. Allopurinol is converted to alloxanthine that inhibits Xanthine Oxidase o Uricosuric drugs increase clearance of uric acid. Probenecid increases renal clearance of uric acid by ihibiting reabsorption Colchecine Anti-Inflammatory— Inhibits microtubule assembly in Leukocytes NSAIDs Anti-Inflammatory Allopurinol Inhibits Xanthine Oxidase Probenecid (Uricosuric) Prevents Uric Acid Reabsorption in the Kidney 6 SCID A deficiency in adenosine Deaminase can Cause Sever Combined Immunodeficiency syndrome Deficit of both T and B cells in the immune system May be due to the build up of dATP which inhibits ribonucleotide Reductase and therefore inhibits DNA synthesis First disease to be successfully treated by gene therapy Purine Nucleoside Phosphorylase Deficiency Deficiency in T-cells, B cells are normal The problem MAY be from an accumulation of dGTP which can inhibit reduction of pyrimidine ribonucleotides by inhibit inhibiting Ribonucleotide Reductase and therefore DNA synthesis Antimetabolites Block metabolic pathways by inhibiting enzyme activity Purine Metabolism blockers o Glutamine Analogs (e.g. Azaserine) Inhibit aminotransferases in purine sythesis Inhibit the CTP sythtetase in pyrimidine synthesis Inhibit enzymes that use glutamine o Purine Nucleotide Analogs Mercaptopurine ihibits phopsphoryl pyrophosphate amidotransferase (PPRP Glutamyl Amidotransferase) Can be used to treat leukemia o Antifolates Folate analogs Inhibits the regeneration of formyl tetrahydrofolate required for steps 4 and 10 of purine synthesis Methotrexate, aminopterin Uses of Antimetabolites o Antimetabolites are widely used in chemotherapy o Some antibiotics are antimetabolites 7 Nucleotides-Pyrimidines, Deoxyribonucleic Acid, and Salvage Pathways 10/15/2008 6:14:00 AM Pyrimidine Biosynthesis Start with simple compounds o Glutamine (N3) o CO2 (C2) o ATP o Aspartate (N1, C6, C5, C4) Glutamine, CO2 and ATP make Carbamoyl Phosphate Aspartate is added All rings are present after the second step PRPP is used, but in the middle end area. PRPP donates the ribose phosphate after ring formation OMP is the first pyrimidine nucleotide then converted to UMP then to UDP o UDP is converted to the other end products. The interconversions must take place at specific phosphorylation levels UDP is phosphorylated to form UTP CTP is converted from UTP by glutamine amino donation TMP is converted from dUMP UDP must undergo a reduction and a dephosphorylation first (Ribonucleotide Reductase then hydrolysis). Enzyme is Thymidylate Synthase The three major end products are CTP, UTP and dTMP Synthesis Pathway 1. Carbamoyl Phosphate Synthetase II i. CO2 + Glutamine glutamate and carbamoyl phosphate ii. Consumes ATP iii. C2 and N3 2. Aspartate Transcarbamoylase i. All members of ring are now present ii. N1, C6, C5, and C4 3. Dihydrotase i. Closes the ring 8 CARBAMOYL PHOSPHATE II , ASPARTATE TRANSCARBAMOYLASE , AND DIHYDROTASE ARE PART OF THE SAME ENZYME (1, 2 and 3) o Referred to as C.A.D. o Use substrate channeling, which helps in regulation and diffusion of substrates from enzyme to ezyme 4. Dihydroorotate Dehydrogenase i. Make the first pyrimidine-Orotic Acid ii. Reduces NAD+ to NADH 5. Orotate Phosphoribosyl Transferase 6. Orotidylic Acid Decarboxylase i. Makes UMP OROTATE PHOSPHORIBOSYL DEHYDROGENASE AND OROTIDYLIC ACID DECARBOXYLASE ARE PART OF THE SAME ENZYME (5 and 6) o Called UMP Synthase 7. UMP is phosphorylated to make UDP. This is where the chain branches. i. UDP is reduced and dephosphorylated (ribonucleotide Reductase) to make dUMP which is then methylated by methylene THF to make dTMP (Thymidylate Synthase). ii. UDP is phosphorylated to make UTP which is then converted to CTP using glutamine as an amino donor (UTPCTP by CTP synthetase) Regulation of Pyrimidine Synthesis End Feedback—CTP inhibits Carbamoyl Phosphate Synthetase II o How does can this only regulate CPII and not CPI? Two ways Compartmentalization—CPI is in the mitochondria and CPII is in the cytosol. There are two distinct pools of Carbamoyl Phosphate. Metabolic Channeling—Carbamoyl Phosphate is not diffusing into cytosol, it is just used right away by Aspartate Transcarbamoylase and then Dihydrotase Inhibition of Pyrimidine Synthesis Glutamine Analogs (Azaserine)—CTP synthetase is inhibited by glutamine analogs by competitive inhibition 9 5-fluorouracil inhibits thymidylate synthase. 5-fluorouracil is converted to 5-fluoro-2-deoxyuridine-5-phosphate (FdUMP) which blocks thymidylate synthase by binding to it irreversibly Catabolism of Pyrimidines Details of the end of the pathway. There are two branches of the pathay that converge Three end products o CO2 o NH3 o -Aminoisobutyrate Can be used to estimate the rate of DNA degradation and cell death. CLINICAL RELEVANCE Hereditary Orotic Aciduria Sxmegaloblastic anemia, growth retardation, excess orotic acid in urine Cause is deficiency in two activities o Orotate phosphoribosyl transferase o Orotidine-5-P decarboxylase Pathophysilogy o Lack of pyrimidines therefore decreased DNA and RNA synthesis o Block in pathway decreases concentration of end feedback mechanism, therefore increased flux through the pathway (until block) Txtreat with Uridine o Kinase will make UTP which will inhibit the pathway and make CTP o Kinase will also make UDP so you can make dTMP Biosynthesis of Deoxyribonucleotides Deoxyribonucleotides are made by reduction of already made diphosphate ribonucleotides Reduction can ONLY OCCUR at the diphosphate level Ribonucleotide Reductase is the enzyme o Uses Thioredoxin to reduce 10 o Three type of sites that can bind nucleotides Catalytic Site-Substrate are any of the ribonucleotides at the diphosphate level Activity Site—allosteric regulator; primarly regulated by the adenosine nucleotides (ATP, dATP) Specifity Site—allosteric regulator; can regulate activity toward other sites dATP binds to the specificity site and that decreases specificity toward all nucleotides ATP binds to specificity site to increase creation of deoxypyrimidines dGTP inhibits pyrimidine deoxyribonucleic acid synthesis and upregulates ADP conversion to dADP dTTP is not made by ribonucleotide reductase but can inhibit it allosterically by binding to the specificity site dCTP does not bind to any allosteric site Why do high levels of thymidine arrest growth of cells? Pools of dTTP, dGTP, and dATP are increased o Reason: dTTP increases action of Ribonucleotide Reductase on GDP which yield dUTP which increases action of Ribonucleotide Reductase on ADP to yield more dATP Pool of dCTP is decreased 10-fold o Reason: dTTP inhibits reduction of pyrimidine nucleoside diphophates by ribonucleotide reductase. dTTP can still be made by salvage pathways, it just cant be made from dUMP Regulation of Ribonucleotide Reductase dATP inhibits at this general activity site dGTP and dTTP act as inhibitors of pyrimidine reduction at the specificity site Synthesis of dTMP Not made from Ribonucleotide Reductase dTMP is made by methylation of dUMP, using methylene-THF s a cofactor 11 Folic Acid Analogs Uses basic structure of analogs, but replaces simple R groups with aminos (and methyls in some cases) Folate Cycle N5N10 Methylene THFFH2 (thymadylate Synthase) o FH2 is not functional FH2 is reduced with NADPH to make THF which then uses serine hydroxymethyl transferase and converts serineglycine o Carbon that is donated comes from SERINE! Woooo o Methotrexate/aminopterin (chemotherapy drugs) block the dihydrofolate Reductase reaction, therefore THF cannot be regenerated How does everything fit together Two different pathways (purine and pyrimidine) The diphosphates will be converted to deoxyribonucleoties o dTTP pathway is long and ridiculous Salvage pathway Recycles bases back to nucleotides Two types o Phosphoribosyl transferase Adds phospho-sugar in one step No enzyme for cytosine (can make from uracil) o Nucleoside phosphorylases and nucleoside kinases Phosphorylase fuses Thymine and deoxyribose1phosphate Kinase phosphoyrlates the nucleoside. CLINICAL RELEVENCE Lesch-Nyhan Syndrome Defect in a phosphoribosyl transferase (HGPRT) Increased uric acid Neurological presentation (brain relies on salvage pathways for nucleotide and is therefore starved for guanine nucleotides Pathphysiology o Lack of HGPRT reduces salvage of guanine and hypoxanthine, thus reducing levels of GMP and IMP. PRPP that is normally 12 used for salvage pathway is used for purine syntheis, leading to incrased degredation and therefore more uric acid 13 Nucleotides-Objectives 10/15/2008 6:14:00 AM 1. Identify the structure of the five common nitrogenous bases found in nucleotides that are incorporated into nucleic acids. Name the nucleosides and nucleotides derived from these bases. 2. Identify the nitrogenous base, sugar, phosphate, glycosidic bond, phosphomonoester bond and phosphoanhydride bonds of a nucleoside triphophate 3. Name the Committed Steps in de novo purine and pyrimidine biosynthesis. a. Purine—PRPP Glutamyl Amidotransferase (Step 2) b. Pyrimidine—CAD; Carbamoyl phosphate synthetase II (Step 1) 4. Compare and contrast the biosynthetic pathways for purines and pyrimidines a. Purine is built upon the sugar where as pyrimidine is added to sugar after is has been made b. Both us PRPP as sourse of ribose 5. Identify the principal metabolic source of one carbon units that are metabolized via tetrahydrofolate a. Serine 6. State the first purine nucleotide that is formed during de novo purine biosynthesis 14 a. Inositol Monophosphate 7. State the first pyrimidine nucleotide that is formed during de novo pyrimidine biosynthesis a. Orotodylic Monophosphate 8. Name the two steps that are regulated early in the de novo purine biosynthetic pathway a. PRPP Synthetase (slightly) b. PRPP Glutamyl Amidotransferase (major) 9. Name the enzyme that is regulated early in the de novo pyrimidine biosynthetic pathway of mammals. a. Caramoyl phosphate synthetase II 10. State the enzyme of nucleotide metabolism that is inhibited by a given antimetabolite a. Glutamine Analogs (Azaserine)—inhibits amidotransferases (rxn 2 and rxn 5 of purine synthesis) b. Purine Nucleotide Analogs (mercaptopurine)—Feed back regulation of PRPP Glutamyl Amidotransferase c. Antifolates (methotrexate, aminopterin)—Inhibit the regeneration of N10-Formyl-THF for Rxn 4 and rxn 10 of purine synthesis and regeneration of N5 N10 methylene THF for Thymidylate synthase by inhibiting Dihydrofolate Reductase (DHFR) d. 5-fluorouracil inhibits Thymidylate Synthetase 11. State the level of phosphorylation at which ribonucleotides are converted to deoxyribonucleotides a. Diphosphate level 12. State which nucleotides allosterically inhibit Ribonucleotide Reductase, and why high levels of thymidine arrest growth of cells in culture 13. Describe the thymidylate synthase reaction and its role in nucleotide biosynthesis. a. Thymidylate synthase reaction 14. Diagram the two preformed or salvage pathways of nucleotide biosynthesis. a. One step i. Adds phophosugar to base 15 b. Two step i. Adds base to sugar ii. Phophorylates sugar 15. State the end product of purine catabolism in man and the precursers that Xanthine Oxidase acts upon to yield this end product a. Uric Acid b. Hypoxanthine and Xanthine 16. State the known enzymatic defects in patients with (and treatments for these diseases (when available) and the mechanism of action of these diseases) a. Gout i. Causes 1. PRPP Synthetase-mutant that’s not regulated will increase nucleotide production and consequently nucleotide degredation 2. Glucose-6-Phosphate Dehydrogenase (leads to increased ribose-5-phosphate, therefore increased nucleotides and furthermore increased nucleotide metabolism) 3. Partial HGPRT deficiency ii. Treatments 1. Anti-inflammatories (NSAIDS-cyclooxygenase, Colchecine-microtuble formation in leukocytes) 2. Allopurinol-Inhibits Xanthine Oxidase 3. Uricosuric (probenecid)-inhibits uric acid reabsorption in the kidney 16 b. Lesch-Nyhan Syndrome i. Defect in HGPRT c. Orotic Aciduria i. d. Xanthinuria i. Xanthine Oxidase deficiency causes Hypoxanthine and Xanthine to be excreted in the urine. Can cause Xanthine Lithiasis in extreme cases. e. Severe Combined Immunodeficiency Syndrome (SCID) i. Cause 1. Adenosine Deaminase deficiency-Build up of dATP which inhibits Ribonucleotide Reductase (makes deoxyribonucleotides from ribonucleotides), thereby inhibiting DNA synthesis. ii. Treatment 1. Gene Therapy-Deliver the gene to the affected tissues and it may be incorporated, thereby fixing the deficiency 17 18